1.9 A structure of the signal receiver domain of the putative response regulator NarL from Mycobacterium tuberculosis - PubMed (original) (raw)
Comparative Study
. 2008 Dec 1;64(Pt 12):1096-100.
doi: 10.1107/S1744309108035203. Epub 2008 Nov 28.
Affiliations
- PMID: 19052358
- PMCID: PMC2593691
- DOI: 10.1107/S1744309108035203
Comparative Study
1.9 A structure of the signal receiver domain of the putative response regulator NarL from Mycobacterium tuberculosis
Robert Schnell et al. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008.
Abstract
NarL from Mycobacterium tuberculosis is a putative nitrate response regulator that is involved in the regulation of anaerobic metabolism in this pathogen. The recombinant purified N-terminal signal receiver domain of NarL has been crystallized in space group C222(1), with unit-cell parameters a = 85.6, b = 90.0, c = 126.3 A, and the structure was determined by molecular replacement to 1.9 A resolution. Comparisons with related signal receiver domains show that the closest structural homologue is an uncharacterized protein from Staphylococcus aureus, whereas the nearest sequence homologue, NarL from Escherichia coli, displays larger differences in three-dimensional structure. The largest differences between the mycobacterial and E. coli NarL domains were found in the loop between beta3 and alpha3 in the proximity of the phosphorylation site. The active site in response regulators is similar to that of members of the haloacid dehalogenase (HAD) family, which also form a phospho-aspartyl intermediate. In NarL, the aspartic acid that acts as catalytic acid/base in several HAD enzymes is replaced by an arginine residue, which is less likely to participate in steps involving proton abstraction. This substitution may slow down the breakdown of the phospho-aspartyl anhydride and allow signalling beyond the timescales defined by a catalytic reaction intermediate.
Figures
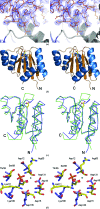
Figure 1
(a) Part of the unbiased 2_F_ o − F c map illustrating the difference density for the fourth molecule (monomer D) in the asymmetric unit. The electron-density map was obtained after locating molecules A, B and C by molecular replacement and refinement using only these three molecules. The refined structure of part of molecule D is included. (b) Cartoon of the overall structure of the signal receiver domain of NarL from M. tuberculosis. Helices are shown in blue and β-strands in brown. The side chain of Asp61, the site of phosphorylation, is indicated as a stick model. (c) Superposition of the Cα traces of the N-terminal signal receiver domains of NarL from M. tuberculosis (blue) and E. coli (green). The side chain of Asp61, the site of phosphorylation, is indicated as a stick model. (d) Comparison of the phosphorylation site in NarL from M. tuberculosis (standard colours) with the active site of histidinol phosphate phosphatase from E. coli (PDB code
2fpw
; blue lines). The active site of the histidinol phosphate phosphatase contains a phosphorylated reaction intermediate, the phosphoryl–aspartic acid mixed anhydride formed at position Asp57, and a Ca2+ ion (indicated by a green sphere) replacing the catalytic Mg2+ ion.
Similar articles
- Mycobacterium tuberculosis response regulators, DevR and NarL, interact in vivo and co-regulate gene expression during aerobic nitrate metabolism.
Malhotra V, Agrawal R, Duncan TR, Saini DK, Clark-Curtiss JE. Malhotra V, et al. J Biol Chem. 2015 Mar 27;290(13):8294-309. doi: 10.1074/jbc.M114.591800. Epub 2015 Feb 6. J Biol Chem. 2015. PMID: 25659431 Free PMC article. - Inhibition of NarL of Mycobacterium Tuberculosis: an in silico approach.
Shivakumar KV, Karunakar P, Chatterjee J. Shivakumar KV, et al. Interdiscip Sci. 2014 Dec;6(4):292-9. doi: 10.1007/s12539-014-0179-z. Epub 2014 Sep 19. Interdiscip Sci. 2014. PMID: 25240609 - Structure of the Escherichia coli response regulator NarL.
Baikalov I, Schröder I, Kaczor-Grzeskowiak M, Grzeskowiak K, Gunsalus RP, Dickerson RE. Baikalov I, et al. Biochemistry. 1996 Aug 27;35(34):11053-61. doi: 10.1021/bi960919o. Biochemistry. 1996. PMID: 8780507 - Serine 83 in DosR, a response regulator from Mycobacterium tuberculosis, promotes its transition from an activated, phosphorylated state to an inactive, unphosphorylated state.
Cho HY, Kang BS. Cho HY, et al. Biochem Biophys Res Commun. 2014 Feb 21;444(4):651-5. doi: 10.1016/j.bbrc.2014.01.128. Epub 2014 Feb 1. Biochem Biophys Res Commun. 2014. PMID: 24491537 - Nitrate regulation of anaerobic respiratory gene expression in Escherichia coli.
Stewart V. Stewart V. Mol Microbiol. 1993 Aug;9(3):425-34. doi: 10.1111/j.1365-2958.1993.tb01704.x. Mol Microbiol. 1993. PMID: 8412692 Review.
Cited by
- The Mycobacterium tuberculosis drugome and its polypharmacological implications.
Kinnings SL, Xie L, Fung KH, Jackson RM, Xie L, Bourne PE. Kinnings SL, et al. PLoS Comput Biol. 2010 Nov 4;6(11):e1000976. doi: 10.1371/journal.pcbi.1000976. PLoS Comput Biol. 2010. PMID: 21079673 Free PMC article. - Preliminary crystallographic studies of the regulatory domain of response regulator YycF from an essential two-component signal transduction system.
Zhao H, Heroux A, Sequeira RD, Tang L. Zhao H, et al. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009 Jul 1;65(Pt 7):719-22. doi: 10.1107/S1744309109022696. Epub 2009 Jun 27. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009. PMID: 19574649 Free PMC article. - Two Fe-S clusters catalyze sulfur insertion by radical-SAM methylthiotransferases.
Forouhar F, Arragain S, Atta M, Gambarelli S, Mouesca JM, Hussain M, Xiao R, Kieffer-Jaquinod S, Seetharaman J, Acton TB, Montelione GT, Mulliez E, Hunt JF, Fontecave M. Forouhar F, et al. Nat Chem Biol. 2013 May;9(5):333-8. doi: 10.1038/nchembio.1229. Epub 2013 Mar 31. Nat Chem Biol. 2013. PMID: 23542644 Free PMC article. - In silico antitubercular activity analysis of benzofuran and naphthofuran derivatives.
Karunakar P, Girija CR, Krishnamurthy V, Krishna V, Shivakumar KV. Karunakar P, et al. Tuberc Res Treat. 2014;2014:697532. doi: 10.1155/2014/697532. Epub 2014 Sep 11. Tuberc Res Treat. 2014. PMID: 25302118 Free PMC article. - A Subgroup of Latently Mycobacterium tuberculosis Infected Individuals Is Characterized by Consistently Elevated IgA Responses to Several Mycobacterial Antigens.
Baumann R, Kaempfer S, Chegou NN, Oehlmann W, Spallek R, Loxton AG, van Helden PD, Black GF, Singh M, Walzl G. Baumann R, et al. Mediators Inflamm. 2015;2015:364758. doi: 10.1155/2015/364758. Epub 2015 Aug 10. Mediators Inflamm. 2015. PMID: 26347586 Free PMC article.
References
- Baikalov, I., Schroder, I., Kaczor-Grzeskowiak, M., Grzeskowiak, K., Gunsalus, R. P. & Dickerson, R. E. (1996). Biochemistry, 35, 11053–11061. - PubMed
- Beier, D. & Gross, R. (2006). Curr. Opin. Microbiol. 9, 143–152. - PubMed
- Birck, C., Mourey, L., Gouet, P., Fabry, B., Schumacher, J., Rousseau, P., Kahn, D. & Samama, J.-P. (1999). Structure, 7, 1505–1515. - PubMed
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials