OPRM1 Asn40Asp predicts response to naltrexone treatment: a haplotype-based approach - PubMed (original) (raw)
OPRM1 Asn40Asp predicts response to naltrexone treatment: a haplotype-based approach
Gabor Oroszi et al. Alcohol Clin Exp Res. 2009 Mar.
Abstract
Background: Individualized pharmacotherapy requires identification of genetic variants predictive of treatment response. In OPRM1, Asn40Asp has been reported to be predictive of response to naltrexone treatment. Nevertheless, the in vitro function of the polymorphism remains elusive and over 300 OPRM1 sequence variants have been identified to date. Therefore we used a haplotype-based approach to capture information of other genetic variants that might predict treatment response to naltrexone in the COMBINE Study.
Methods: 5' nuclease genotyping assays (TaqMan) were applied for 10 SNPs. Five-locus haplotypes in 2 OPRM1 haplotype blocks were assigned to Caucasian participants. The relationship of the haplotypes to medication reflected by "good clinical outcome" was analyzed in 306 Caucasians treated without Combined Behavioral Intervention and with either naltrexone or placebo.
Results: A significant haplotype by medication interaction (p = 0.03) was found in OPRM1 block 1. Naltrexone-treated alcoholics with haplotype AGCCC, the single haplotype carrying the Asp40 allele had the highest percent of good clinical outcome. When interaction of genotypes at each of the 5 loci comprising block 1 with medication was examined, only the Asn40/Asp40 and Asp40/Asp40 genotypes were found to significantly interact with naltrexone treatment. No haplotype by medication interaction was documented in OPRM1 block 2.
Conclusions: Our haplotype-based approach confirms that the single OPRM1 locus predictive of response to naltrexone treatment is Asn40Asp in exon 1. A substantial contribution of any other OPRM1 genetic variant to interindividual variations in response to naltrexone treatment (at least in terms of good clinical outcome) is not supported by our findings.
Conflict of interest statement
Disclosure/Conflict of Interest
Dr. Anton has reported receiving consultation fees and honoraria from Forest Laboratories and Alkermes (the maker of long-acting injectable naltrexone); consultation fees and grants from Bristol-Myers Squibb and Hythiam; Consultation fees, honoraria, and grants from Contral Pharma/Biotie Pharmaceuticals and Johnson & Johnson/OrthoMcNeil; consultation fees and grant funding from Pfizer; and, consultation fees from AstraZeneca, Axis Sheild, Cephalon, Drug Abuse Sciences, and Sanofi Aventis. In the near future he anticipates receiving consulting fees from Solvay Pharmaceuticals and a grant from Eli Lilly. Dr. O’Malley has reported receiving research support (grant support or clinical supplies) from Alkermes, DuPont, GlaxoSmithKline, Forest Laboratories, Lipha Pharmaceuticals, Ortho-McNeil, Bristol-Myers Squibb, Pfizer, Sanofi-Aventis, and Mallinckrodt and anticipates receiving a contract from Eli Lilly; serving as a consultant to Alkermes, Forest Laboratories, GlaxoSmithKline, Ortho-McNeil, Pfizer, Johnson & Johnson; receiving travel reimbursement from Alkermes; and that she is an inventor on patents held by Yale University entitled: “Smoking Cessation Treatments Using Naltrexone and Related Compounds”. Dr. Pettinati has reported receiving research support from Alkermes, AstraZeneca, Bristol-Myers Squibb, Cephalon, Forest Laboratories, Lipha-Merck-KGaA, Ortho- McNeil; and, serving as consultant/advisory board/speakers bureau for Alkermes, AstraZeneca, Cephalon, and Forest Laboratories. Dr. Swift has reported receiving grant funding from Ortho-McNeill and Pfizer, Inc.; serving as consultant/advisory board/speakers bureau to Alkermes, Inc., Cephalon, Ortho-McNeil, Pfizer, Inc., Transoral Pharmaceuticals, Forest Laboratories. Drs. Oroszi, Couper, Yuan and Goldman had nothing to disclose.
Figures
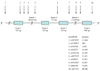
Figure 1
Schematic (non-scaled) structure of the human μ opioid receptor gene (OPRM1) based on the MOR-1 transcript variant (NM_000914.2). Positions of 10 SNPs genotyped and their dbSNP IDs are also shown. Nucleotide +1 is the A of the ATG translation initiation codon, the nucleotide 5’ to +1 is numbered −1.
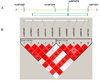
Figure 2
(A) Scaled schematic structure of OPRM1 (on line version is in color). The yellow (top) bar represents the contig sequence spanning 20kb upstream and 10kb downstream of the 5’ and 3’ ends of the gene indicated by the green (lower) rectangle. The green vertical bars in the rectangle indicate exons. (B) Linkage disequilibrium (LD) plot of 10 OPRM1 SNPs based on 685 Caucasians in the COMBINE Study. The D’ value of each SNP pair is shown in the squares. The numbers in the squares are D’x 100. Empty squares indicate D’ = 1. Squares are colored bright red (dark grey) if the D’ value is high and the confidence in the value of D’ is high as well. The first and the last markers in each block are also displayed on the structure of OPRM1 to help the comparison between the two parts of the figure.
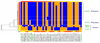
Figure 3
Close correspondence of haplotypes in the COMBINE Study and HapMap and haplotype grouping (on line version is in color). Alleles of markers (SNPs) genotyped in both datasets are highlighted in green (bold). The first allele and second allele in the haplotypes are depicted in blue (dark grey) and brown (light grey), respectively. The height of each haplotype is proportional to the CEU haplotype frequency in HapMap. Haplotypes with a frequency ≥ 3% are shown in both datasets. a). Block 1: The SNP (rs1799971) for Asn40Asp(A118G) is highlighted in bold italics b). Block 2.
Figure 3
Close correspondence of haplotypes in the COMBINE Study and HapMap and haplotype grouping (on line version is in color). Alleles of markers (SNPs) genotyped in both datasets are highlighted in green (bold). The first allele and second allele in the haplotypes are depicted in blue (dark grey) and brown (light grey), respectively. The height of each haplotype is proportional to the CEU haplotype frequency in HapMap. Haplotypes with a frequency ≥ 3% are shown in both datasets. a). Block 1: The SNP (rs1799971) for Asn40Asp(A118G) is highlighted in bold italics b). Block 2.
Similar articles
- An evaluation of mu-opioid receptor (OPRM1) as a predictor of naltrexone response in the treatment of alcohol dependence: results from the Combined Pharmacotherapies and Behavioral Interventions for Alcohol Dependence (COMBINE) study.
Anton RF, Oroszi G, O'Malley S, Couper D, Swift R, Pettinati H, Goldman D. Anton RF, et al. Arch Gen Psychiatry. 2008 Feb;65(2):135-44. doi: 10.1001/archpsyc.65.2.135. Arch Gen Psychiatry. 2008. PMID: 18250251 Free PMC article. Clinical Trial. - Naltrexone modification of drinking effects in a subacute treatment and bar-lab paradigm: influence of OPRM1 and dopamine transporter (SLC6A3) genes.
Anton RF, Voronin KK, Randall PK, Myrick H, Tiffany A. Anton RF, et al. Alcohol Clin Exp Res. 2012 Nov;36(11):2000-7. doi: 10.1111/j.1530-0277.2012.01807.x. Epub 2012 May 2. Alcohol Clin Exp Res. 2012. PMID: 22551036 Free PMC article. Clinical Trial. - Pharmacogenetics of naltrexone in asian americans: a randomized placebo-controlled laboratory study.
Ray LA, Bujarski S, Chin PF, Miotto K. Ray LA, et al. Neuropsychopharmacology. 2012 Jan;37(2):445-55. doi: 10.1038/npp.2011.192. Epub 2011 Sep 7. Neuropsychopharmacology. 2012. PMID: 21900886 Free PMC article. Clinical Trial. - The μ-opioid receptor and treatment response to naltrexone.
Thorsell A. Thorsell A. Alcohol Alcohol. 2013 Jul-Aug;48(4):402-8. doi: 10.1093/alcalc/agt030. Epub 2013 Mar 29. Alcohol Alcohol. 2013. PMID: 23543091 Review. - The role of the Asn40Asp polymorphism of the mu opioid receptor gene (OPRM1) on alcoholism etiology and treatment: a critical review.
Ray LA, Barr CS, Blendy JA, Oslin D, Goldman D, Anton RF. Ray LA, et al. Alcohol Clin Exp Res. 2012 Mar;36(3):385-94. doi: 10.1111/j.1530-0277.2011.01633.x. Epub 2011 Sep 6. Alcohol Clin Exp Res. 2012. PMID: 21895723 Free PMC article. Review.
Cited by
- Neural correlates of adherence to extended-release naltrexone pharmacotherapy in heroin dependence.
Wang AL, Elman I, Lowen SB, Blady SJ, Lynch KG, Hyatt JM, O'Brien CP, Langleben DD. Wang AL, et al. Transl Psychiatry. 2015 Mar 17;5(3):e531. doi: 10.1038/tp.2015.20. Transl Psychiatry. 2015. PMID: 25781230 Free PMC article. - An initial investigation of associations between dopamine-linked genetic variation and smoking motives in African Americans.
Bidwell LC, McGeary JE, Gray JC, Palmer RH, Knopik VS, MacKillop J. Bidwell LC, et al. Pharmacol Biochem Behav. 2015 Nov;138:104-10. doi: 10.1016/j.pbb.2015.09.018. Epub 2015 Sep 26. Pharmacol Biochem Behav. 2015. PMID: 26410615 Free PMC article. - Pharmacotherapies and personalized medicine for alcohol use disorder: a review.
Lohoff FW. Lohoff FW. Pharmacogenomics. 2020 Oct;21(15):1117-1138. doi: 10.2217/pgs-2020-0079. Epub 2020 Aug 18. Pharmacogenomics. 2020. PMID: 32807012 Free PMC article. Review. - Haplotype block structure of the genomic region of the mu opioid receptor gene.
Levran O, Awolesi O, Linzy S, Adelson M, Kreek MJ. Levran O, et al. J Hum Genet. 2011 Feb;56(2):147-55. doi: 10.1038/jhg.2010.150. Epub 2010 Dec 16. J Hum Genet. 2011. PMID: 21160491 Free PMC article. - Systematic Review of Polygenic Gene-Environment Interaction in Tobacco, Alcohol, and Cannabis Use.
Pasman JA, Verweij KJH, Vink JM. Pasman JA, et al. Behav Genet. 2019 Jul;49(4):349-365. doi: 10.1007/s10519-019-09958-7. Epub 2019 May 20. Behav Genet. 2019. PMID: 31111357 Free PMC article.
References
- Anton RF, O'Malley SS, Ciraulo DA, Cisler RA, Couper D, Donovan DM, Gastfriend DR, Hosking JD, Johnson BA, LoCastro JS, Longabaugh R, Mason BJ, Mattson ME, Miller WR, Pettinati HM, Randall CL, Swift R, Weiss RD, Williams LD, Zweben A. Combined pharmacotherapies and behavioral interventions for alcohol dependence: the COMBINE study: a randomized controlled trial. JAMA. 2006;295(17):2003–2017. - PubMed
- Anton RF, Oroszi G, O'Malley S, Couper D, Swift R, Pettinati H, Goldman D. An evaluation of mu-opioid receptor (OPRM1) as a predictor of naltrexone response in the treatment of alcohol dependence: results from the Combined Pharmacotherapies and Behavioral Interventions for Alcohol Dependence (COMBINE) study. Arch Gen Psychiatry. 2008;65(2):135–144. - PMC - PubMed
- Arias A, Feinn R, Kranzler HR. Association of an Asn40Asp (A118G) polymorphism in the mu-opioid receptor gene with substance dependence: a meta-analysis. Drug Alcohol Depend. 2006;83(3):262–268. - PubMed
- Bare LA, Mansson E, Yang D. Expression of two variants of the human mu opioid receptor mRNA in SK-N-SH cells and human brain. FEBS Lett. 1994;354(2):213–216. - PubMed
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U10AA11776/AA/NIAAA NIH HHS/United States
- U10AA11777/AA/NIAAA NIH HHS/United States
- Z99 AA999999/ImNIH/Intramural NIH HHS/United States
- U10 AA011715/AA/NIAAA NIH HHS/United States
- U10AA11721/AA/NIAAA NIH HHS/United States
- U10AA11773/AA/NIAAA NIH HHS/United States
- U10AA11799/AA/NIAAA NIH HHS/United States
- U10AA11787/AA/NIAAA NIH HHS/United States
- U10AA11756/AA/NIAAA NIH HHS/United States
- U10AA11768/AA/NIAAA NIH HHS/United States
- U10AA11715/AA/NIAAA NIH HHS/United States
- U10 AA011715-03/AA/NIAAA NIH HHS/United States
- Z01 AA000305-04/ImNIH/Intramural NIH HHS/United States
- U10AA11783/AA/NIAAA NIH HHS/United States
- U10AA11727/AA/NIAAA NIH HHS/United States
- U10AA11716/AA/NIAAA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical