Genome stability is ensured by temporal control of kinetochore-microtubule dynamics - PubMed (original) (raw)
Genome stability is ensured by temporal control of kinetochore-microtubule dynamics
Samuel F Bakhoum et al. Nat Cell Biol. 2009 Jan.
Abstract
Most solid tumours are aneuploid and many frequently mis-segregate chromosomes. This chromosomal instability is commonly caused by persistent mal-oriented attachment of chromosomes to spindle microtubules. Chromosome segregation requires stable microtubule attachment at kinetochores, yet those attachments must be sufficiently dynamic to permit correction of mal-orientations. How this balance is achieved is unknown, and the permissible boundaries of attachment stability versus dynamics essential for genome stability remain poorly understood. Here we show that two microtubule-depolymerizing kinesins, Kif2b and MCAK, stimulate kinetochore-microtubule dynamics during distinct phases of mitosis to correct mal-orientations. Few-fold reductions in kinetochore-microtubule turnover, particularly in early mitosis, induce severe chromosome segregation defects. In addition, we show that stimulation of microtubule dynamics at kinetochores restores stability to chromosomally unstable tumour cell lines, establishing a causal relationship between deregulation of kinetochore-microtubule dynamics and chromosomal instability. Thus, temporal control of microtubule attachment to chromosomes during mitosis is central to genome stability in human cells.
Figures
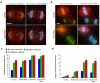
Figure 1. Lagging chromosomes in the absence of Kif2b and MCAK
a, Anaphase spindles of both untreated and Kif2b-deficient U2OS cells containing CenpB-GFP to indicate kinetochores and stained for microtubules and DNA. White arrow highlights merotelic kinetochores. For clarity, a cell with only 2 lagging chromosomes was selected for this panel. b, Anaphase and pre-anaphase cells of both untreated and Kif2b-deficient RPE1 cells following recovery from nocodazole treatment stained for kinetochores with CREST, cyclin B, and DNA. c, d, Number of anaphase U2OS cells with lagging kinetochores (c) and the average number of laggards per anaphase (d) in untreated cells (blue) or cells following recovery from monastrol (green) or nocodazole (red) treatment. Bars represent mean ± s.e.m, n = 150 cells, 3 experiments. Scale bars, 5 μm.
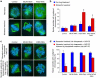
Figure 2. Kif2b is required for correction of attachment errors
a, Spindles of untreated (Control), MCAK-deficient (MCAK RNAi), and Kif2b-deficient (Kif2b RNAi) U2OS cells in the presence of monastrol (top panels) or after recovery from monastrol treatment (bottom panels). b, Number of cells with bipolar spindles that failed to align chromosomes with no treatment (blue) or following recovery from monastrol treatment (red). c, Spindles of untreated (Control), MCAK-deficient (MCAK RNAi), and Kif2b-deficient (Kif2b RNAi) cells following recovery from monastrol treatment in the presence of hesperadin (top panels) and then followed by recovery from hesperadin treatment (bottom panels). d, Number of bipolar cells that failed to align chromosomes after monastrol washout in the presence of hesperadin (blue) and then after recovery from hesperadin treatment (red). Bars in b and d represent mean ± s.e.m, n = 600, 3 experiments cells for b, 100 cells, 2 experiments for d. Scale bars, 5 μm. *, p < 0.0002, t-test.
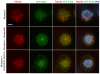
Figure 3. Kinetochore localization of GFP-Kif2b is sensitive to the Aurora-inhibitor, Hesperadin
Monopolar spindles induced with monastrol showing microtubules (red), GFP-Kif2b (green), and DNA (blue) in the presence of hesperadin and following removal of hesperadin. Scale bar, 5 μm.
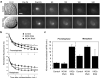
Figure 4. Temporal regulation of kinetochore-microtubule dynamics
a, Differential interference contrast (D.I.C.) and time-lapse fluorescent images of prometaphase spindles in untreated (Control) and Kif2b-deficient (Kif2b RNAi) U2OS cells before (Pre-PA) and at the indicated times (sec) after activation (Post-PA) of GFP-tubulin fluorescence. Scale bar, 5 μm. b, Normalized fluorescence intensity over time after photoactivation of spindles in untreated (filled squares), MCAK-deficient (filled circles), and Kif2b-deficient (white squares) prometaphase and metaphase cells. Datapoints represent mean ± s.e.m, n = 5 to 11 cells. c, Calculated kMT half-life under different conditions. Error bars represent S.E. from the regression analysis in b.
Figure 5. Suppression of chromosome mis-segregation in cancer cell lines
a, b, Percent of anaphase cells with lagging kinetochores and average numbers of lagging chromosomes in anaphase of U2OS cells from representative experiments. Clone 5 and clone 8 are from Table 1. Bars represent mean ± s.e.m, n = 150 cells, 3 experiments (a) and bars represent values from 100 cells, 2 experiements (b), *, p < 0.05, t-test. **c,** Examples of a proper chromosome segregation in U2OS cells expressing GFP-MCAK (left panel) and mis-segregation event in U2OS cells expressing GFP-Kif2a (right panel) using FISH with probes for chromosomes 2 (green) and 3 (red) and DNA stained with DAPI (blue). Scale bars, 10 μm. **d,** Mis-segregation rates per chromosome per mitosis in untreated U2OS cells and U2OS cells overexpressing GFP-Kif2a, GFP-Kif2b, GFP-MCAK, or GFP-tubulin as indicated. Clone 5 and clone 8 are from Table 1. n > 660 cells. *, p < 0.05, Chi-square test. e, Model for temporal regulation of kMT dynamics. At low inter-kinetochore tension during prometaphase, the Aurora B kinase activity gradient recruits Kif2b to kinetochores and inhibits centromeric MCAK. The tension generated upon biorientation causes sister kinetochores to exceed the boundaries of the Aurora B kinase activity gradient releasing Kif2b from kinetochores and activating a subset of MCAK in the outer centromere. Kinetochores are shown in grey and microtubules are shown in green.
Similar articles
- CLASP1, astrin and Kif2b form a molecular switch that regulates kinetochore-microtubule dynamics to promote mitotic progression and fidelity.
Manning AL, Bakhoum SF, Maffini S, Correia-Melo C, Maiato H, Compton DA. Manning AL, et al. EMBO J. 2010 Oct 20;29(20):3531-43. doi: 10.1038/emboj.2010.230. Epub 2010 Sep 17. EMBO J. 2010. PMID: 20852589 Free PMC article. - The dynamic kinetochore-microtubule interface.
Maiato H, DeLuca J, Salmon ED, Earnshaw WC. Maiato H, et al. J Cell Sci. 2004 Nov 1;117(Pt 23):5461-77. doi: 10.1242/jcs.01536. J Cell Sci. 2004. PMID: 15509863 Review. - BubR1 and CENP-E have antagonistic effects upon the stability of microtubule-kinetochore attachments in Drosophila S2 cell mitosis.
Maia AF, Lopes CS, Sunkel CE. Maia AF, et al. Cell Cycle. 2007 Jun 1;6(11):1367-78. doi: 10.4161/cc.6.11.4271. Epub 2007 Jun 11. Cell Cycle. 2007. PMID: 17525528 - Merotelic kinetochore orientation, aneuploidy, and cancer.
Cimini D. Cimini D. Biochim Biophys Acta. 2008 Sep;1786(1):32-40. doi: 10.1016/j.bbcan.2008.05.003. Epub 2008 May 23. Biochim Biophys Acta. 2008. PMID: 18549824 Review. - Microtubule flux: what is it good for?
Khodjakov A, Kapoor T. Khodjakov A, et al. Curr Biol. 2005 Dec 6;15(23):R966-8. doi: 10.1016/j.cub.2005.11.017. Curr Biol. 2005. PMID: 16332529 Review.
Cited by
- Spatiotemporal dynamics of Aurora B-PLK1-MCAK signaling axis orchestrates kinetochore bi-orientation and faithful chromosome segregation.
Shao H, Huang Y, Zhang L, Yuan K, Chu Y, Dou Z, Jin C, Garcia-Barrio M, Liu X, Yao X. Shao H, et al. Sci Rep. 2015 Jul 24;5:12204. doi: 10.1038/srep12204. Sci Rep. 2015. PMID: 26206521 Free PMC article. - Centromeres are dismantled by foundational meiotic proteins Spo11 and Rec8.
Hou H, Kyriacou E, Thadani R, Klutstein M, Chapman JH, Cooper JP. Hou H, et al. Nature. 2021 Mar;591(7851):671-676. doi: 10.1038/s41586-021-03279-8. Epub 2021 Mar 3. Nature. 2021. PMID: 33658710 Free PMC article. - Chromosome instability in neuroblastoma: A pathway to aggressive disease.
Paolini L, Hussain S, Galardy PJ. Paolini L, et al. Front Oncol. 2022 Oct 20;12:988972. doi: 10.3389/fonc.2022.988972. eCollection 2022. Front Oncol. 2022. PMID: 36338721 Free PMC article. Review. - Accurate chromosome segregation by probabilistic self-organisation.
Saka Y, Giuraniuc CV, Ohkura H. Saka Y, et al. BMC Biol. 2015 Aug 12;13:65. doi: 10.1186/s12915-015-0172-y. BMC Biol. 2015. PMID: 26264961 Free PMC article. - Polo regulates Spindly to prevent premature stabilization of kinetochore-microtubule attachments.
Barbosa J, Martins T, Bange T, Tao L, Conde C, Sunkel C. Barbosa J, et al. EMBO J. 2020 Jan 15;39(2):e100789. doi: 10.15252/embj.2018100789. Epub 2019 Dec 18. EMBO J. 2020. PMID: 31849090 Free PMC article.
References
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. - PubMed
- Storchova Z, Pellman D. From polyploidy to aneuploidy, genome instability and cancer. Nat. Rev. Mol. Cell Biol. 2004;5:45–54. - PubMed
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancer. Nature. 1997;386:623–627. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA023108/CA/NCI NIH HHS/United States
- GM008704/GM/NIGMS NIH HHS/United States
- R01 GM051542-12/GM/NIGMS NIH HHS/United States
- GM51542/GM/NIGMS NIH HHS/United States
- R37 GM051542/GM/NIGMS NIH HHS/United States
- R01 GM051542/GM/NIGMS NIH HHS/United States
- T32 GM008704/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials