Microsporidia are natural intracellular parasites of the nematode Caenorhabditis elegans - PubMed (original) (raw)
Microsporidia are natural intracellular parasites of the nematode Caenorhabditis elegans
Emily R Troemel et al. PLoS Biol. 2008.
Abstract
For decades the soil nematode Caenorhabditis elegans has been an important model system for biology, but little is known about its natural ecology. Recently, C. elegans has become the focus of studies of innate immunity and several pathogens have been shown to cause lethal intestinal infections in C. elegans. However none of these pathogens has been shown to invade nematode intestinal cells, and no pathogen has been isolated from wild-caught C. elegans. Here we describe an intracellular pathogen isolated from wild-caught C. elegans that we show is a new species of microsporidia. Microsporidia comprise a large class of eukaryotic intracellular parasites that are medically and agriculturally important, but poorly understood. We show that microsporidian infection of the C. elegans intestine proceeds through distinct stages and is transmitted horizontally. Disruption of a conserved cytoskeletal structure in the intestine called the terminal web correlates with the release of microsporidian spores from infected cells, and appears to be part of a novel mechanism by which intracellular pathogens exit from infected cells. Unlike in bacterial intestinal infections, the p38 MAPK and insulin/insulin-like growth factor (IGF) signaling pathways do not appear to play substantial roles in resistance to microsporidian infection in C. elegans. We found microsporidia in multiple wild-caught isolates of Caenorhabditis nematodes from diverse geographic locations. These results indicate that microsporidia are common parasites of C. elegans in the wild. In addition, the interaction between C. elegans and its natural microsporidian parasites provides a system in which to dissect intracellular intestinal infection in vivo and insight into the diversity of pathogenic mechanisms used by intracellular microbes.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
Figure 1. Intracellular Infection of C. elegans Proceeds through Distinct Stages
(A) Uninfected intestine. (B) Infection causes displacement of gut granules, which appear as “grooves,” indicated by arrow. (C) Small rod-shaped microbes in intestine, indicated by arrow. Arrowheads indicate intestinal lumen in (A–C). (D) Higher magnification view of rod-shaped microbes. Asterisk marks gut granule. (E) Large and small rod-shaped microbes are indicated by larger and smaller arrow, respectively. (F) Vesicles of microbes are indicated by arrow. Scale bar is 10 μm in (A–C), (E), (F), and 2 μm in (D).
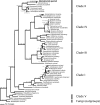
Figure 2. Phylogenetic Analysis of Microsporidian Sequence Isolated from Infected Nematodes
Bayesian inference phylogeny of the small subunit ribosomal DNA from selected microsporidia and close relatives, estimated using the program MrBayes. Two fungi were used as outgroups in the analysis (see [29]). Numbers above branches are Bayesian posterior probabilities/parsimony bootstrap/maximum likelihood bootstrap values. If nodal support values were below 0.5 or below 50% for any analysis we indicated this by “—“. The five major clades of Microsporidia are indicated on the right of the figure (see [29]). Note the relationship between Nematocida and O. popilliae.
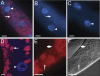
Figure 3. Intracellular Microbe Is a Novel Microsporidia Species
(A–E) FISH staining of C. elegans intestines using Cy3 probes for rRNA in red and (A–D) counterstained with DAPI in blue to label DNA. (A) Infected animal at the groove stage stained with MicroA, a probe specific for N. parisii rRNA. Arrow indicates FISH and DAPI staining of multinucleate microbes (meronts). Similar results were obtained with MicroB, another probe specific for N. parisii (unpublished data). (B) Infected animal at the groove stage stained with universal probe for bacterial rRNA (EUB338) [67]. No FISH staining is observed, although excess DAPI staining is visible due to meronts, indicated with arrow. (C) Uninfected animal stained with N. parisii-specific probe. For (A–C) scale bar is 10 μm. (D) Confocal imaging of FISH staining with _N. parisii-_specific probe. Arrow indicates multinucleate microbes (meronts). Scale bar is 5 μm. For (A and D), arrowhead indicates host nucleus. (E) rRNA FISH staining of rod-shaped microbes (spores) with N. parisii-specific probe. (F) DIC image of spores. For (E and F), small spores indicated with smaller arrow, large spores indicated by larger arrow, and scale bar is 10 μm.
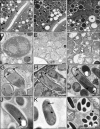
Figure 4. Transmission Electron Micrographs of N. parisii Infection in the C. elegans Intestine
(A) Uninfected adult intestine. (B) Adult intestine 30 h postinoculation (at groove stage by light microscopy). N. parisii meronts are indicated with arrows. In (A and B), double arrows indicate microvilli and Lu indicates lumen. Scale bar is 2 μm. (C) Higher magnification view of multinucleate meront indicated with an arrow in close proximity to host vesicle marked with an asterisk. Scale bar is 1 μm. (D) Later stage meront with two nuclei and more regularly shaped plasma membrane, 44 h postinoculation. (E and F) Adult intestine 44 h postinoculation with stages intermediate between meront and spore. (G–I) Adult intestine 44 h postinoculation containing developing N. parisii spores. (J) Mature spore. (K) Larger sized spore (comparable to larger microbes in Figure 1E). For (G–K) PT refers to the anterior portion of the polar tube called the manubroid [68], PP refers to the polaroplast membranes, PV refers to the posterior vacuole and arrowheads indicate cross-sections of polar tube coils. (L) Vesicles surrounding spores (comparable to vesicles in Figure 1F). For (D–L), scale bar is 500 nm.
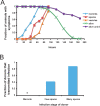
Figure 5. Timecourse of N. parisii Infection and Transmission of Infection
(A) Progression of infection in wild-type animals. L1s were infected with N. parisii at 0 h, and 80–150 animals were scored at each timepoint for meronts, spores, or vesicles by DIC microscopy, and for survival by prodding animals with a platinum wire. “Alive uninf.” refers to survival of uninfected animals grown in parallel with infected animals. The time to 50% of animals exhibiting symptoms was 43.1 ± 3.7 h for meronts, 65.8 h for spores, and 75.9 ± 1.6 h for vesicles. The time to 50% animals dead (TD50) was 127 ± 2 h. There was a statistically significant difference between the survival of infected and uninfected animals from 116 h onward (116 h, p = .02; 138.5 h, p = .0057; 163.5 h, p = .00015; 186.5 h, p = .00011). See Materials and Methods for analysis details. (B) Transmission of infection. Fraction of infected donor animals that transmitted infection is indicated on _y_-axis. None of the 16 donors infected only with meronts transmitted the infection (asterisk indicates fraction equals 0). 16 of 18 donors with many spores (>100 spores) transmitted the infection. Three of seven donors with only a few spores (<100 spores) transmitted the infection. See Materials and Methods for assay details.
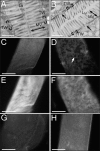
Figure 6. Infected Animals Have Lesions in the Terminal Web
(A) Electron micrograph showing cross-section of intestine in uninfected animal. (B) Micrograph showing cross-section of intestine in N. parisii infected animal. Gaps in terminal web are indicated with arrows. (A, B) Brackets indicate terminal web (TW), double arrow indicates microvilli (MV), Lu indicates lumen, scale bar is 1 μm. (C) Uninfected intestine stained with MH33 antibody, which labels IFB-2, an intermediate filament component in the terminal web. Terminal web staining appears as a sheet, since image is from nonconfocal microscope. (D) Infected intestine stained with MH33. Gaps in MH33 staining are indicated with an arrow. (E) Same uninfected intestine as in (C), but visualizing ERM-1::GFP. (F) Same infected intestine as in (D), but visualizing ERM-1::GFP. Animal stained with MH33 antibody after being infected with P. aeruginosa strain PA14 for 27 h (G) or S. aureus strain NCTC 8325 for 24 h (H). Note gut distension due to PA14 infection. Scale bar is 10 μm (C–H).
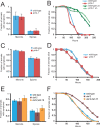
Figure 7. N. parisii Infection in Wild-Type and pmk-1 Mutant Animals
(A and C) Comparison of infection symptoms in wild-type and pmk-1 mutants infected with N. parisii at the L1 stage and assayed at 43 h postinoculation (A) or infected at the L4 stage and then assayed 47 h postinoculation (C), with at least 40 animals per plate. The average of three plates is shown, error bars are standard deviation. There was not a significant difference between wild-type and pmk-1 at the L1 stage (p = 0.75 for meronts, p = 0.39 for spores) or the L4 stage (p = 0.10 for meronts, p = 0.53 for spores). (B and D) Comparison of survival between wild-type and pmk-1(km25) mutants inoculated with N. parisii at L1 stage (B) or L4 stage (D). Assays were performed starting with at least 40 animals per plate, three plates per experiment. In (D), “un” refers to uninfected. There was not a significant difference between wild-type and pmk-1 at the L1 stage (p = 0.0529) or the L4 stage (p = 0.65). (E) Comparison of infection symptoms in wild-type, daf-2(e1368), and daf-2(e1368);daf-16(mgDf47) mutants infected with N. parisii at the L4 stage and assayed at 45 h postinoculation. The average of three plates is shown, error bars are standard deviation. There was not a significant difference between wild-type and daf-2 (p = 0.69 for meronts, p = 0.44 for spores), between wild-type and daf-2;daf-16 (p = 0.15 for meronts, p = 0.22 for spores) or between daf-2 and daf-2;daf-16 (p = 0.19 for meronts, p = 0.17 for spores). (F) Comparison of survival between wild-type, daf-2(e1368), and daf-2(e1368);daf-16(mgDf47) mutants infected with N. parisii at the L4 stage. Assays were performed starting with at least 40 animals per plate, three plates per experiment. There was a significant difference between survival of wild-type and daf-2 (p < 0.0001) and between daf-2 and daf-2;daf-16 (p < 0.0001), but not between wild-type and daf-2;daf-16 (p = 0.40). All data shown in the figure are representative of at least three independent experiments.
Figure 8. Microsporidia-Infected Caenorhabditis Nematodes Isolated from Diverse Geographical Locations
(A) Infected dauer larva isolated in Franconville, France. (B) Infected C. elegans adult isolated in Santeuil, France. This adult was filled with rod-shaped microbes and unable to lay eggs. Progeny inside the adult are indicated with asterisks. (C) Infected L2 larva isolated in Lisbon, Portugal. This individual (like some of the other infected larvae) died before reaching adulthood and producing progeny. (D) Infected L4 larva isolated in Santeuil, France. This animal also died before producing progeny. DIC micrographs taken within 24 h after sampling for (A–D). (E) Infected F1 progeny of C. elegans isolated in Montsoreau, France. (F) Infected progeny of C. briggsae from India. (G–I) RNA FISH staining with MicroA, a probe for N. parisii rRNA. (G) JU1247, an infected C. elegans strain isolated in Santeuil, France. (H) JU1395, an infected C. elegans strain isolated in Montsoreau, France. (I) JU1348, an infected C. briggsae strain isolated in Kerala, India. (A–I) Smaller arrow indicates smaller rod-shaped microbes, larger arrow indicates larger rod-shaped microbes, both of which are microsporidian spores.
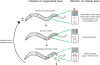
Figure 9. Model of N. parisii Infection in C. elegans on Cellular and Organismal Level
See text for details.
Similar articles
- A Large Collection of Novel Nematode-Infecting Microsporidia and Their Diverse Interactions with Caenorhabditis elegans and Other Related Nematodes.
Zhang G, Sachse M, Prevost MC, Luallen RJ, Troemel ER, Félix MA. Zhang G, et al. PLoS Pathog. 2016 Dec 12;12(12):e1006093. doi: 10.1371/journal.ppat.1006093. eCollection 2016 Dec. PLoS Pathog. 2016. PMID: 27942022 Free PMC article. - Host-Microsporidia Interactions in Caenorhabditis elegans, a Model Nematode Host.
Troemel ER. Troemel ER. Microbiol Spectr. 2016 Oct;4(5). doi: 10.1128/microbiolspec.FUNK-0003-2016. Microbiol Spectr. 2016. PMID: 27763260 Review. - A new microsporidium Percutemincola moriokae gen. nov., sp. nov. from Oscheius tipulae: A novel model of microsporidia-nematode associations.
Nishikori K, Setiamarga DHE, Tanji T, Kuroda E, Shiraishi H, Ohashi-Kobayashi A. Nishikori K, et al. Parasitology. 2018 Dec;145(14):1853-1864. doi: 10.1017/S0031182018000628. Epub 2018 Apr 17. Parasitology. 2018. PMID: 29661263 - Cell-to-cell spread of microsporidia causes Caenorhabditis elegans organs to form syncytia.
Balla KM, Luallen RJ, Bakowski MA, Troemel ER. Balla KM, et al. Nat Microbiol. 2016 Aug 22;1(11):16144. doi: 10.1038/nmicrobiol.2016.144. Nat Microbiol. 2016. PMID: 27782144 Free PMC article. - Microsporidia: Pervasive natural pathogens of Caenorhabditis elegans and related nematodes.
Gang SS, Lažetić V. Gang SS, et al. J Eukaryot Microbiol. 2024 Sep-Oct;71(5):e13027. doi: 10.1111/jeu.13027. Epub 2024 May 3. J Eukaryot Microbiol. 2024. PMID: 38702921 Review.
Cited by
- Specific detection and localization of microsporidian parasites in invertebrate hosts by using in situ hybridization.
Dubuffet A, Smith JE, Solter L, Perotti MA, Braig HR, Dunn AM. Dubuffet A, et al. Appl Environ Microbiol. 2013 Jan;79(1):385-8. doi: 10.1128/AEM.02699-12. Epub 2012 Oct 19. Appl Environ Microbiol. 2013. PMID: 23087031 Free PMC article. - Conservation of Nematocida microsporidia gene expression and host response in Caenorhabditis nematodes.
Wan YC, Troemel ER, Reinke AW. Wan YC, et al. PLoS One. 2022 Dec 19;17(12):e0279103. doi: 10.1371/journal.pone.0279103. eCollection 2022. PLoS One. 2022. PMID: 36534656 Free PMC article. - Microsporidia-nematode associations in methane seeps reveal basal fungal parasitism in the deep sea.
Sapir A, Dillman AR, Connon SA, Grupe BM, Ingels J, Mundo-Ocampo M, Levin LA, Baldwin JG, Orphan VJ, Sternberg PW. Sapir A, et al. Front Microbiol. 2014 Feb 10;5:43. doi: 10.3389/fmicb.2014.00043. eCollection 2014. Front Microbiol. 2014. PMID: 24575084 Free PMC article. - Transcriptomic profiling of host-parasite interactions in the microsporidian Trachipleistophora hominis.
Watson AK, Williams TA, Williams BA, Moore KA, Hirt RP, Embley TM. Watson AK, et al. BMC Genomics. 2015 Nov 21;16:983. doi: 10.1186/s12864-015-1989-z. BMC Genomics. 2015. PMID: 26589282 Free PMC article. - Fungal Diversity in Tomato Rhizosphere Soil under Conventional and Desert Farming Systems.
Kazerooni EA, Maharachchikumbura SSN, Rethinasamy V, Al-Mahrouqi H, Al-Sadi AM. Kazerooni EA, et al. Front Microbiol. 2017 Aug 2;8:1462. doi: 10.3389/fmicb.2017.01462. eCollection 2017. Front Microbiol. 2017. PMID: 28824590 Free PMC article.
References
- Bhavsar AP, Guttman JA, Finlay BB. Manipulation of host-cell pathways by bacterial pathogens. Nature. 2007;449:827–834. - PubMed
- Cossart P, Sansonetti PJ. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science. 2004;304:242–248. - PubMed
- Kazmierczak BI, Mostov K, Engel JN. Interaction of bacterial pathogens with polarized epithelium. Annu Rev Microbiol. 2001;55:407–435. - PubMed
- Hamon M, Bierne H, Cossart P. Listeria monocytogenes: a multifaceted model. Nat Rev Microbiol. 2006;4:423–434. - PubMed
Publication types
MeSH terms
Grants and funding
- P30 DK040561/DK/NIDDK NIH HHS/United States
- P30 DK043351/DK/NIDDK NIH HHS/United States
- F32 AI069732/AI/NIAID NIH HHS/United States
- DK57521/DK/NIDDK NIH HHS/United States
- F32AI069732/AI/NIAID NIH HHS/United States
- R01 AI064332/AI/NIAID NIH HHS/United States
- P01 AI044220/AI/NIAID NIH HHS/United States
- DK43351/DK/NIDDK NIH HHS/United States
- R01 AI072508/AI/NIAID NIH HHS/United States
- P30 DK057521/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases