PASTAA: identifying transcription factors associated with sets of co-regulated genes - PubMed (original) (raw)
PASTAA: identifying transcription factors associated with sets of co-regulated genes
Helge G Roider et al. Bioinformatics. 2009.
Abstract
Motivation: A major challenge in regulatory genomics is the identification of associations between functional categories of genes (e.g. tissues, metabolic pathways) and their regulating transcription factors (TFs). While, for a limited number of categories, the regulating TFs are already known, still for many functional categories the responsible factors remain to be elucidated.
Results: We put forward a novel method (PASTAA) for detecting transcriptions factors associated with functional categories, which utilizes the prediction of binding affinities of a TF to promoters. This binding strength information is compared to the likelihood of membership of the corresponding genes in the functional category under study. Coherence between the two ranked datasets is seen as an indicator of association between a TF and the category. PASTAA is applied primarily to the determination of TFs driving tissue-specific expression. We show that PASTAA is capable of recovering many TFs acting tissue specifically and, in addition, provides novel associations so far not detected by alternative methods. The application of PASTAA to detect TFs involved in the regulation of tissue-specific gene expression revealed a remarkable number of experimentally supported associations. The validated success for various datasets implies that PASTAA can directly be applied for the detection of TFs associated with newly derived gene sets.
Availability: The PASTAA source code as well as a corresponding web interface is freely available at http://trap.molgen.mpg.de.
Figures
Fig. 1.
The PASTAA workflow.
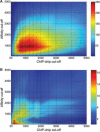
Fig. 2.
Cut-off space for the hypergeometric test. (A) The −log hypergeometric _P_-values (indicated by colour) for ABF1_01 and the Abf1 in vitro dataset depending on the cut-off combination employed for the predicted affinity and PBM binding values. The most significant target enrichment (_P_-value 7.3 × 10−253) is found when using the top 800 genes according to PBM and top 900 genes according to affinity. The steepest increase in −log _P_-values is found at the origin of the plot. (B) Same analysis as in (A) but for the factor PHO4_01 and the Pho4 ChIP–chip dataset (phosphate-deprived condition). According to the fact that Pho4 has far less targets than Abf1 an optimal hypergeometric _P_-value of 7.9 × 10−20 is found when using only the top 300 genes according to ChIP–chip data and top 100 genes according to affinity.
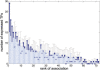
Fig. 3.
TFs are over-expressed in their top ranking tissues. Height of bins indicates the number of TFs expressed in the associated tissue of given rank based on the real sequence data (dark blue) or on the results obtained from 10 random sequence sets (light blue). Error bars show the 95% confidence interval for the results obtained from the 10 random sequence sets. Tissues top ranking for a given TF express the factor more often than expected, while bottom ranking tissues express the TF equally or less often than expected. The enrichment is particularly significant for the first three bins corresponding to all three top ranking TF–tissue associations (_P_-value of enrichment for bins 1–3 combined: 2.2 × 10−12). The general trend in the light blue bins indicates the technical bias caused by the different number of ESTs in each tissue category.
Similar articles
- Condition-specific target prediction from motifs and expression.
Meng G, Vingron M. Meng G, et al. Bioinformatics. 2014 Jun 15;30(12):1643-50. doi: 10.1093/bioinformatics/btu066. Epub 2014 Feb 14. Bioinformatics. 2014. PMID: 24532727 - CORE_TF: a user-friendly interface to identify evolutionary conserved transcription factor binding sites in sets of co-regulated genes.
Hestand MS, van Galen M, Villerius MP, van Ommen GJ, den Dunnen JT, 't Hoen PA. Hestand MS, et al. BMC Bioinformatics. 2008 Nov 26;9:495. doi: 10.1186/1471-2105-9-495. BMC Bioinformatics. 2008. PMID: 19036135 Free PMC article. - Comparative promoter region analysis powered by CORG.
Dieterich C, Grossmann S, Tanzer A, Röpcke S, Arndt PF, Stadler PF, Vingron M. Dieterich C, et al. BMC Genomics. 2005 Feb 21;6:24. doi: 10.1186/1471-2164-6-24. BMC Genomics. 2005. PMID: 15723697 Free PMC article. - KnockTF: a comprehensive human gene expression profile database with knockdown/knockout of transcription factors.
Feng C, Song C, Liu Y, Qian F, Gao Y, Ning Z, Wang Q, Jiang Y, Li Y, Li M, Chen J, Zhang J, Li C. Feng C, et al. Nucleic Acids Res. 2020 Jan 8;48(D1):D93-D100. doi: 10.1093/nar/gkz881. Nucleic Acids Res. 2020. PMID: 31598675 Free PMC article. - Network motif-based identification of transcription factor-target gene relationships by integrating multi-source biological data.
Zhang Y, Xuan J, de los Reyes BG, Clarke R, Ressom HW. Zhang Y, et al. BMC Bioinformatics. 2008 Apr 21;9:203. doi: 10.1186/1471-2105-9-203. BMC Bioinformatics. 2008. PMID: 18426580 Free PMC article.
Cited by
- The NSL complex regulates housekeeping genes in Drosophila.
Lam KC, Mühlpfordt F, Vaquerizas JM, Raja SJ, Holz H, Luscombe NM, Manke T, Akhtar A. Lam KC, et al. PLoS Genet. 2012;8(6):e1002736. doi: 10.1371/journal.pgen.1002736. Epub 2012 Jun 14. PLoS Genet. 2012. PMID: 22723752 Free PMC article. - Inferring direct DNA binding from ChIP-seq.
Bailey TL, Machanick P. Bailey TL, et al. Nucleic Acids Res. 2012 Sep 1;40(17):e128. doi: 10.1093/nar/gks433. Epub 2012 May 18. Nucleic Acids Res. 2012. PMID: 22610855 Free PMC article. - Patterns of transcription factor binding and epigenome at promoters allow interpretable predictability of multiple functions of non-coding and coding genes.
Chandra O, Sharma M, Pandey N, Jha IP, Mishra S, Kong SL, Kumar V. Chandra O, et al. Comput Struct Biotechnol J. 2023 Jul 14;21:3590-3603. doi: 10.1016/j.csbj.2023.07.014. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37520281 Free PMC article. - In silico discovery of novel transcription factors regulated by mTOR-pathway activities.
Jablonska A, Polouliakh N. Jablonska A, et al. Front Cell Dev Biol. 2014 Jun 3;2:23. doi: 10.3389/fcell.2014.00023. eCollection 2014. Front Cell Dev Biol. 2014. PMID: 25364730 Free PMC article. - E2F1 controls germ cell apoptosis during the first wave of spermatogenesis.
Rotgers E, Nurmio M, Pietilä E, Cisneros-Montalvo S, Toppari J. Rotgers E, et al. Andrology. 2015 Sep;3(5):1000-14. doi: 10.1111/andr.12090. Andrology. 2015. PMID: 26311345 Free PMC article.
References
- Aoki KF, Kanehisa M. Unit 1. 12: Using the KEGG database resource, Chapter 1. Curr. Protoc. Bioinformatics. 2005 - PubMed
- Bailey TL, Elkan C. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1995;3:21–29. - PubMed
- Carninci P, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous