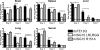ISG15 Arg151 and the ISG15-conjugating enzyme UbE1L are important for innate immune control of Sindbis virus - PubMed (original) (raw)
ISG15 Arg151 and the ISG15-conjugating enzyme UbE1L are important for innate immune control of Sindbis virus
Nadia V Giannakopoulos et al. J Virol. 2009 Feb.
Abstract
Interferon (IFN)-stimulated gene 15 (ISG15) is a ubiquitin-like molecule that conjugates to target proteins via a C-terminal LRLRGG motif and has antiviral function in vivo. We used structural modeling to predict human ISG15 (hISG15) residues important for interacting with its E1 enzyme, UbE1L. Kinetic analysis revealed that mutation of arginine 153 to alanine (R153A) ablated hISG15-hUbE1L binding and transthiolation of UbcH8. Mutation of other predicted UbE1L-interacting residues had minimal effects on the transfer of ISG15 from UbE1L to UbcH8. The capacity of hISG15 R153A to form protein conjugates in 293T cells was markedly diminished. Mutation of the homologous residue in mouse ISG15 (mISG15), arginine 151, to alanine (R151A) also attenuated protein ISGylation following transfection into 293T cells. We assessed the role of ISG15-UbE1L interactions in control of virus infection by constructing double subgenomic Sindbis viruses that expressed the mISG15 R151A mutant. While expression of mISG15 protected alpha/beta-IFN-receptor-deficient (IFN-alphabetaR(-/-)) mice from lethality following Sindbis virus infection, expression of mISG15 R151A conferred no survival benefit. The R151A mutation also attenuated ISG15's ability to decrease Sindbis virus replication in IFN-alphabetaR(-/-) mice or prolong survival of ISG15(-/-) mice. The importance of UbE1L was confirmed by demonstrating that mice lacking this ISG15 E1 enzyme were highly susceptible to Sindbis virus infection. Together, these data support a role for protein conjugation in the antiviral effects of ISG15.
Figures
FIG. 1.
Mutation of UbE1L-interacting residue arginine 153 in hISG15 affects protein conjugate formation. (A) Western blot analysis of six-His hISG15 (GG) and six-His hISG15 UbE1L-interacting mutant R153A following cotransfection with hUbE1L, UbcH8, and Herc5 into 293T cells. Asterisks indicate nonspecific bands detected in mock-transfected samples, and numbers (for panels A, C, D, and E) represent molecular mass markers (kDa). (B) Alignment of hISG15 and mISG15 sequences. Highlighted amino acids indicate ISG15 residues predicted to interact with UbE1L from hISG15 crystallization studies (33). R153A and R151A indicate hISG15 and mISG15 residues mutated in this study. (C and D) Anti-mISG15 Western blot analysis of 293T cells transfected with mISG15 LRLRGG (GG), conjugation-deficient mISG15 LRLRAA (AA), and UbE1L-interacting mutant mISG15 R151A (R151A) in the absence (C) or presence (D) of an E3 ligase, Herc5. (E) Representative anti-mISG15 Western blot analysis of IFN-β-treated ISG15−/− MEFs transfected with mISG15 LRLRGG (GG), mISG15 LRLRAA (AA), or mISG15 R151A (R151A) (top panel). The blot was reprobed for GFP (bottom panel). (F) Quantization of Western blot signals in panel E. Error bars represent the standard errors of the means from four experiments. One-sample t tests indicated that both mISG15 LRLRAA and mISG15 R151A (asterisks) were significantly different from mISG15 LRLRGG (mISG15 LRLRAA, P = 0.0015; 95% confidence interval, −0.264 to 4.294; mISG15 R151A, P = 0.0045; 95% confidence interval, 0.876 to 6.199). The graph is an arbitrary scale with mISG15 LRLRGG assigned a value of 10.
FIG. 2.
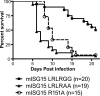
Mutation of UbE1L-interacting residue Arg151 attenuates mISG15's antiviral function during Sindbis virus infection. (A) Western blot analysis of BHK-21 cells mock infected (M) or infected with parental dsTE12Q (Q) or dsTE12Q containing mISG15 LRLRGG (GG), mISG15 LRLRAA (AA), or mISG15 R151A (R151A) at an MOI of 20. Parallel blots were probed with anti-ISG15 (top panel) or polyclonal anti-Sindbis virus (bottom panel). (B) Single-step growth curves of parental (dsTE12Q) or recombinant (mISG15 LRLRGG, mISG15 LRLRAA, or mISG15 R151A) Sindbis viruses in BHK-21 cells (MOI, 5). Data are represented as means ± standard errors of the means for three (dsTE12Q) or six (recombinant viruses) replicates. There are no significant differences between the medians of different viruses when analyzed by analysis of variance (P = 0.9993). (C) Survival of IFN-αβR−/− mice infected with 5 × 106 PFU of Sindbis viruses s.c. as indicated. The number of mice infected per group is indicated in parentheses, and data are compiled from a minimum of three experiments. Asterisks indicate curves statistically different from mISG15 LRLRGG (LRLRGG versus R151A, LRLRGG versus LRLRAA, and LRLRGG versus dsTE12Q, all P < 0.001).
FIG. 3.
ISG15 UbE1L-interacting mutant R151A is attenuated in its ability to decrease Sindbis virus replication. Viral titers of dsTE12Q, mISG15 LRLRGG, or mISG15 R151A in brains, spleen, liver, lung, and serum of IFN-αβR−/− mice infected for 3, 5, or 7 days. Data are pooled from two independent experiments with six mice per group. Error bars represent standard errors of the means, and the dashed line indicates the plaque assay limit of detection. Brackets denote statistically significant (P < 0.05) differences between indicated viruses.
FIG. 4.
mISG15 UbE1L-interacting mutants prolong survival during Sindbis virus infection. Survival of ISG15−/− pups infected with 1,000 PFU intracerebrally of virus expressing either mISG15 LRLRGG, mISG15 LRLRAA, or mISG15 R151A. Data are pooled from two (mISG15 R151A) or three (mISG15 LRLRGG and mISG15 LRLRAA) experiments, and the numbers of mice per group are indicated in parentheses. P values for comparisons between survival curves are as follows: mISG15 R151A versus mISG15 LRLRGG, P = 0.0002; mISG15 R151A versus mISG15 LRLRAA, P = 0.0365; mISG15 LRLRGG versus mISG15 LRLRAA, P < 0.0001.
FIG. 5.
UbE1L−/− mice display increased susceptibility to Sindbis virus infection. (A) Anti-mISG15 Western blot analysis of brains from BL6 or UbE1L−/− mice infected with 5 × 106 PFU of dsTE12Q s.c. on day 3 or 7 postinfection. Parallel blots were probed with anti-mISG15 (top panel) or anti-β-actin (bottom panel). DPI, day postinfection. (B) Survival of UbE1L−/− and BL6 mice following infection with 5 × 106 PFU of dsTE12Q s.c. P values of log-rank comparisons between UbE1L−/− and BL6 mouse survival curves are shown.
Similar articles
- The basis for selective E1-E2 interactions in the ISG15 conjugation system.
Durfee LA, Kelley ML, Huibregtse JM. Durfee LA, et al. J Biol Chem. 2008 Aug 29;283(35):23895-902. doi: 10.1074/jbc.M804069200. Epub 2008 Jun 26. J Biol Chem. 2008. PMID: 18583345 Free PMC article. - Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo.
Lenschow DJ, Giannakopoulos NV, Gunn LJ, Johnston C, O'Guin AK, Schmidt RE, Levine B, Virgin HW 4th. Lenschow DJ, et al. J Virol. 2005 Nov;79(22):13974-83. doi: 10.1128/JVI.79.22.13974-13983.2005. J Virol. 2005. PMID: 16254333 Free PMC article. - Up-regulation of interferon-stimulated gene 15 and its conjugation machinery, UbE1L and UbcH8 expression by tumor necrosis factor-α through p38 MAPK and JNK signaling pathways in human lung carcinoma.
Lertsooksawat W, Wongnoppavich A, Chairatvit K. Lertsooksawat W, et al. Mol Cell Biochem. 2019 Dec;462(1-2):51-59. doi: 10.1007/s11010-019-03609-5. Epub 2019 Aug 19. Mol Cell Biochem. 2019. PMID: 31428903 - ISG15 and immune diseases.
Jeon YJ, Yoo HM, Chung CH. Jeon YJ, et al. Biochim Biophys Acta. 2010 May;1802(5):485-96. doi: 10.1016/j.bbadis.2010.02.006. Epub 2010 Feb 12. Biochim Biophys Acta. 2010. PMID: 20153823 Free PMC article. Review. - The ISG15/USP18 ubiquitin-like pathway (ISGylation system) in hepatitis C virus infection and resistance to interferon therapy.
Chen L, Li S, McGilvray I. Chen L, et al. Int J Biochem Cell Biol. 2011 Oct;43(10):1427-31. doi: 10.1016/j.biocel.2011.06.006. Epub 2011 Jun 16. Int J Biochem Cell Biol. 2011. PMID: 21704181 Review.
Cited by
- Viral hijacking of the host ubiquitin system to evade interferon responses.
Viswanathan K, Früh K, DeFilippis V. Viswanathan K, et al. Curr Opin Microbiol. 2010 Aug;13(4):517-23. doi: 10.1016/j.mib.2010.05.012. Epub 2010 Jun 17. Curr Opin Microbiol. 2010. PMID: 20699190 Free PMC article. Review. - ISG15 conjugation system targets the viral NS1 protein in influenza A virus-infected cells.
Zhao C, Hsiang TY, Kuo RL, Krug RM. Zhao C, et al. Proc Natl Acad Sci U S A. 2010 Feb 2;107(5):2253-8. doi: 10.1073/pnas.0909144107. Epub 2010 Jan 19. Proc Natl Acad Sci U S A. 2010. PMID: 20133869 Free PMC article. - UBE2L6/UBCH8 and ISG15 attenuate autophagy in esophageal cancer cells.
Falvey CM, O'Donovan TR, El-Mashed S, Nyhan MJ, O'Reilly S, McKenna SL. Falvey CM, et al. Oncotarget. 2017 Apr 4;8(14):23479-23491. doi: 10.18632/oncotarget.15182. Oncotarget. 2017. PMID: 28186990 Free PMC article. - ISGylation of NF-κBp65 by SCFFBXL19 E3 Ligase Diminishes Endothelial Inflammation.
Li L, Miao J, Shaheen N, Taleb SJ, Hu J, Ye Q, He J, Yan J, Mallampalli RK, Zhao J, Zhao Y. Li L, et al. Arterioscler Thromb Vasc Biol. 2023 May;43(5):674-683. doi: 10.1161/ATVBAHA.122.318894. Epub 2023 Mar 30. Arterioscler Thromb Vasc Biol. 2023. PMID: 36994728 Free PMC article. - The interferon-induced gene ISG15 blocks retrovirus release from cells late in the budding process.
Pincetic A, Kuang Z, Seo EJ, Leis J. Pincetic A, et al. J Virol. 2010 May;84(9):4725-36. doi: 10.1128/JVI.02478-09. Epub 2010 Feb 17. J Virol. 2010. PMID: 20164219 Free PMC article.
References
- Abraham, N., D. F. Stojdl, P. I. Duncan, N. Methot, T. Ishii, M. Dube, B. C. Vanderhyden, H. L. Atkins, D. A. Gray, M. W. McBurney, A. E. Koromilas, E. G. Brown, N. Sonenberg, and J. C. Bell. 1999. Characterization of transgenic mice with targeted disruption of the catalytic domain of the double-stranded RNA-dependent protein kinase, PKR. J. Biol. Chem. 2745953-5962. - PubMed
- Blomstrom, D. C., D. Fahey, R. Kutny, B. D. Korant, and E. Knight, Jr. 1986. Molecular characterization of the interferon-induced 15-kDa protein. Molecular cloning and nucleotide and amino acid sequence. J. Biol. Chem. 2618811-8816. - PubMed
- Bohnsack, R. N., and A. L. Haas. 2003. Conservation in the mechanism of Nedd8 activation by the human AppBp1-Uba3 heterodimer. J. Biol. Chem. 27826823-26830. - PubMed
- Dastur, A., S. Beaudenon, M. Kelley, R. M. Krug, and J. M. Huibregtse. 2006. Herc5, an interferon-induced HECT E3 enzyme, is required for conjugation of ISG15 in human cells. J. Biol. Chem. 2814334-4338. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI080672/AI/NIAID NIH HHS/United States
- R01 GM047426/GM/NIGMS NIH HHS/United States
- GM47426/GM/NIGMS NIH HHS/United States
- GM34007/GM/NIGMS NIH HHS/United States
- U54 AI057160/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous