The multiple roles of cohesin in meiotic chromosome morphogenesis and pairing - PubMed (original) (raw)
The multiple roles of cohesin in meiotic chromosome morphogenesis and pairing
Gloria A Brar et al. Mol Biol Cell. 2009 Feb.
Abstract
Sister chromatid cohesion, mediated by cohesin complexes, is laid down during DNA replication and is essential for the accurate segregation of chromosomes. Previous studies indicated that, in addition to their cohesion function, cohesins are essential for completion of recombination, pairing, meiotic chromosome axis formation, and assembly of the synaptonemal complex (SC). Using mutants in the cohesin subunit Rec8, in which phosphorylated residues were mutated to alanines, we show that cohesin phosphorylation is not only important for cohesin removal, but that cohesin's meiotic prophase functions are distinct from each other. We find pairing and SC formation to be dependent on Rec8, but independent of the presence of a sister chromatid and hence sister chromatid cohesion. We identified mutations in REC8 that differentially affect Rec8's cohesion, pairing, recombination, chromosome axis and SC assembly function. These findings define Rec8 as a key determinant of meiotic chromosome morphogenesis and a central player in multiple meiotic events.
Figures
Figure 1.
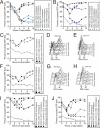
An assay to monitor pairing in live cells. (A) Wild-type cells with homologous LYS2 dots (A9828, ■), wild-type cells with nonhomologous LYS2/URA3 dots (A9829, ○), and wild-type cells with nonhomologous LEU2/CEN5 dots (A16360, ▵), all deleted for NDT80, were introduced into sporulation medium. At the indicated times, samples were taken and assayed for pairing live on aliquots that are taken at the indicated times as cells progress through prophase. Paired GFP signals are not distinguishable, and only a single GFP dot is visible. Nonhomologous arrays are included in each experiment as a control for clustering of the tet operators. Note that the same nonhomologous dot controls are used in A–E as the data from these panels were generated from the same experiment. (B) Wild-type cells with homologous LEU2 dots (A5111, ■), wild-type cells with nonhomologous LYS2/URA3 dots (A9829, ○), and wild-type cells with nonhomologous LEU2/CEN5 dots (A16360, ▵), all deleted for NDT80, were assayed for pairing as described in A. (C) Wild-type cells with homologous URA3 dots (A6946, ■), wild-type cells with nonhomologous LYS2/URA3 dots (A9829, ○), and wild-type cells with nonhomologous LEU2/CEN5 dots (A16360, ▵), all deleted for NDT80, were assayed for pairing as described in A. (D) Wild-type cells with homologous CEN5 dots (A16362, ■), wild-type cells with nonhomologous LYS2/URA3 dots (A9829, ○), and wild-type cells with nonhomologous LEU2/CEN5 dots (A16360, ▵), all deleted for NDT80, were assayed for pairing as described in A. (E) Wild-type cells with homologous TEL5 dots (A16366, ■), wild-type cells with nonhomologous LYS2/URA3 dots (A9829, ○) and wild-type cells with nonhomologous LEU2/CEN5 dots (A16360, ▵), all deleted for NDT80, were assayed for pairing as described in A. (F) Wild-type cells with homologous LYS2 dots (A9828, ■), _spo11_Δ cells with homologous LYS2 dots (A11469, ▴), spo11-Y135F cells with homologous LYS2 dots (A12095, ·), and wild-type cells with nonhomologous LYS2/URA3 dots (A9829, □), all deleted for NDT80, were assayed for pairing as described in Figure (A). Note that the same nonhomologous dot control strain is used in F and G as the data from these panels were generated from the same experiment. (G) Wild-type cells with homologous URA3 dots (A6946, ■), _spo11_Δ cells with homologous URA3 dots (A7803, ▴), spo11-Y135F cells with homologous URA3 dots (A9115, ·), and wild-type cells with nonhomologous LYS2/URA3 dots (A9829, □), all deleted for NDT80, were assayed for pairing as described in A. (H) Wild-type cells with homologous LEU2 dots (A5111, ■), _dmc1_Δ cells with homologous LEU2 dots (A6917, ▴), and wild-type cells with nonhomologous LEU2/LYS2 dots (A11474, □), all deleted for NDT80, were assayed for pairing as described in A. (I) Wild-type cells with homologous LYS2 dots (A9828, ■), _msh5_Δ cells with homologous LYS2 dots (A16290, ▴) and wild-type cells with nonhomologous URA3/LYS2 dots (A9829, □), all deleted for NDT80, were assayed for pairing as described in A.
Figure 2.
REC8 but not sister chromatid cohesion is required for pairing. (A) Wild-type cells with homologous TEL5 dots (A16366, ■), or with homologous URA3 dots (A6946, ▴), or with homologous CEN5 dots (A16362, ·), _rec8_Δ cells with homologous TEL5 dots (A16535, blue rectangle), or with homologous URA3 dots, (A16538, blue triangles), or with homologous CEN5 dots (A16537, blue circles), and wild-type cells with nonhomologous CEN5/LEU2 dots (A16360, ○), all deleted for NDT80, were assayed for pairing as described in Figure 1A. (B) Wild-type cells with homologous LYS2 dots (A9828, ■), or with homologous LEU2 dots (A5111, ▴), _rec8_Δ cells with homologous LYS2 dots (A16108, blue rectangles), or with homologous LEU2 dots, (A16131, blue triangles), and wild-type cells with nonhomologous LYS2/LEU2 dots (A11474, □), all deleted for NDT80, were assayed for pairing as described in Figure 1A. (C–E) Wild-type (A9828, ■) or cdc6-mn cells with homologous LYS2 dots (A10735, ▴) and wild-type cells with nonhomologous URA3/LYS2 dots (A9829, □), all deleted for NDT80, were induced to sporulate to assayed pairing as described in Figure 1A (C) or DNA content by flow cytometry analysis (D and E). (F–H) Wild-type cells with homologous URA3 dots (A6946, ■), cdc6-mn cells with homologous URA3 dots (A10404, ▴), wild-type cells with nonhomologous URA3/LYS2 dots (A9829, □), all deleted for NDT80, were induced to sporulate to assayed pairing as described in Figure 1A (F) or DNA content by flow cytometry analysis (G and H). (I) Wild-type cells with homologous LEU2 dots (A5111, ■), _rec8_Δ cells with homologous LEU2 dots (A16131, ▴), cdc6-mn cells with homologous LEU2 dots (A16533, ·), _rec8_Δ cdc6-mn cells with homologous LEU2 dots (A16460, ♦), and wild-type cells with nonhomologous LEU2/LYS2 dots (A11474, □), all deleted for NDT80, were assayed for pairing as described in Figure 1A. Note that the time courses shown in I and J were performed at the same time, and the nonhomologous dot controls are shown in both experiments. (J) Wild-type cells with homologous LYS2 dots (A9828, ■), _rec8_Δ cells with homologous LYS2 dots (A16108, ▴), cdc6-mn cells with homologous LYS2 dots (A10735, ·), _rec8_Δ cdc6-mn cells with homologous LYS2 dots (A16446, ♦), and wild-type cells with nonhomologous LEU2/LYS2 dots (A11474, □), all deleted for NDT80, were assayed for pairing as described in Figure 1A.
Figure 3.
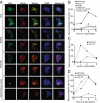
Meiotic cohesin complexes are required for Zip1 assembly. (A) Examples of meiotic cells that were harvested and assayed for Zip1 staining on chromosome spreads. Cells carry a Rec8-3HA construct. α-Zip1 staining is shown in green, α-HA in red, and DNA in blue. (B) Wild-type (A7097, ■), _spo11_Δ (A8477, ▴), _rec8_Δ (A16664, ·), cdc6-mn (A15880, □), _cdc6-mn rec8_Δ (A17021, ▵), and _clb5_Δ _clb6_Δ (A16113, ○) were induced to sporulate. At the indicated times, cells were harvested and chromosome spreads were assayed for Zip1 staining. In this and subsequent experiments, the “percentage of mononucleates with Zip1” encompasses cells with partially and fully assembled Zip1 as defined in A. The percentage of cells in the individual Zip1 categories is shown in Supplemental Figure S4. The meiotic progression of these strains is shown in Supplemental Figure S3A. In this and subsequent experiments 100 mononucleate cells were counted per strain per time point. (C) Wild-type (A1972, ■) and scc3-mn (A20163, ·) were induced to sporulate. At the indicated times, cells were harvested and chromosome spreads were assayed for Zip1 staining. The percentage of cells in the individual Zip1 categories is shown in Supplemental Figure S5. The meiotic progression of these strains is shown in Supplemental Figure S3B. (D) REC8-3HA (A13946, ■), pREC8-SCC1-3HA (A16132, ▴), and REC8-NC (A13539, ·) were induced to sporulate. At the indicated times, cells were harvested and chromosome spreads were assayed for Zip1 staining. The percentage of cells in the individual Zip1 categories is shown in Supplemental Figure S6. The meiotic progression of these strains is shown in Supplemental Figure S3C.
Figure 4.
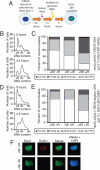
Postreplicative Rec8 is sufficient for Zip1 assembly in the presence of DSBs. (A) The experimental scheme used in B–E. _rec8::pGAL1-REC8-3HA spo11_Δ _rec8_Δ GAL4-ER cells were induced to sporulate, allowed 4.5 h to progress through meiosis, and then treated with 1 μM β-estradiol (+βE). After another 1.5 h incubation in sporulation medium, cells were γ-irradiated with 20KRad (+IR) to induce DSBs. Cells were then kept in sporulation medium for another 3 h, when they were harvested and assayed for α-Zip1 and α-HA staining. A sample was also taken at 6.5 h to assay for DSBs by α-Rad51 staining. (B and C) _rec8::pGAL1-REC8-3HA spo11_Δ _rec8_Δ _GAL4-ER ndt80_Δ (A19800) cells were induced to sporulate and treated as described in A. At 9 h, cells with the indicated treatments were harvested, and chromosome spreads were assayed for Zip1 staining (C). DNA content was determined by flow cytometry analysis of cells harvested at 0 and 4.5 h of sporulation (B). (D and E) _rec8::pGAL1-REC8-3HA-degron spo11_Δ _GAL4-ER ndt80_Δ (A19798) cells were induced to sporulate and treated as described in A. At 9 h, cells with the indicated treatments were harvested, and chromosome spreads were assayed for Zip1 staining (E). DNA content was determined by flow cytometry analysis of cells harvested at 0 and 4.5 h of sporulation (D). (F) Examples of Rec8 and Rad51 staining in cells treated as described in A. At 6.5 h, cells were harvested and chromosome spreads were stained for Rec8, Rad51, and DNA. α-HA is shown in green, α-Rad51 in red, and DNA in blue.
Figure 5.
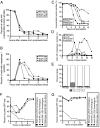
rec8-6A and rec8-29A cells form sister chromatid cohesion. (A and B) Wild-type (A21230, ■), rec8-17A (A21234, ▴), and rec8-29A (21232, ·), all carrying GAL-NDT80 and GAL4-ER fusions, were induced to sporulate in the absence of βE. βE was added after 6 h, and samples were taken to determine the percentage of cells with unassembled meiotic spindles (A) and of cells with metaphase I spindles (B). n = 200 cells counted per time point per strain. (C and D) Wild-type (A1972, ■), _rec8_Δ (A3528, ▴), rec8-29A (A14385, ·), and rec8-6A (A15042, □) cells were induced to sporulate. At the indicated times, samples were taken to determine the percentage of cells with unassembled spindles (A) and of cells with metaphase I spindles (B). n = 200 cells counted per strain per time point. (E) _spo11_Δ _spo13_Δ (A21466, A21467, and A21468), _spo11_Δ _spo13_Δ _rec8_Δ (A21472, A21473, and A21474), _spo11_Δ _spo13_Δ rec8-29A (A21469, A21470, and A21471), and _spo11_Δ _spo13_Δ rec8-6A (A21463, A21464, and A21465) cells carrying a tet repressor-GFP fusion construct and one heterozygous tandem tet operator array inserted at the LEU2 locus were induced to sporulate on plates. After 24 h of sporulation, dyads were scored for sister chromatid segregation by fluorescence microscopy. Cells in which sister chromatids properly segregated apart are represented by the white portion of the bar, whereas cells in which sister chromatids segregate together are represented by the gray portion of the bar. n = 100 cells counted per strain in three independent isogenic diploid strains. Error bars, SD. (F) Wild-type cells with homologous LYS2 dots (A9828, ■), rec8-6A cells with homologous LYS2 dots (A21669, ▴, rec8-29A cells with homologous LYS2 dots (A16412, ·), and wild-type cells with nonhomologous LEU2/LYS2 dots (A11474, □), all deleted for NDT80, assayed for pairing as described in Figure 1A. Note that the same nonhomologous dot control strain is shown in F and G, because the experiments were performed in parallel. (G) Wild-type cells with homologous LEU2 dots (A5111, ■), rec8-6A cells with homologous LEU2 dots (A21670, ▴), rec8-29A cells with homologous LEU2 dots (A21668, ·), and wild-type cells with nonhomologous LEU2/LYS2 dots (A11474, □), all deleted for NDT80, were assayed for pairing as described in Figure 1A.
Figure 6.
rec8-29A mutant cells exhibit defects in homologous recombination. (A) Wild-type (A1556), _rec8_Δ (A18933), and _pREC8-SCC1 rec8_Δ (A16132) were induced to sporulate. At the indicated times, cells were harvested and assayed by Southern blot for DSBs and recombination products at HIS4/LEU2. (B) Wild-type (A1556), _rec8_Δ (A18933), rec8-29A (A21618), and rec8-6A (A18936) cells were induced to sporulate. At the indicated times, cells were harvested and assayed by Southern blot for DSBs and recombination products at HIS4/LEU2. (C and D) Blots from A and B were subjected to densitometric analysis to quantitate the intensity of the bands representing the upper recombinant band. Values were normalized to the “Mom” parental band for each strain and each time point. (E) Meiotic progression of the strains assayed for recombination shown in A and B. Wild-type (A1556, ◇), _rec8_Δ (A18933, ■), _pREC8-SCC1 rec8_Δ (A16132, ○), rec8-29A (A21618, ·), and rec8-6A (A18936, ▴) cells were induced to sporulate. At the indicated times, samples were taken and subjected to α-tubulin immunofluorescence to determine the percentage of cells with unassembled spindles. n = 200 cells counted per strain per time point.
Figure 7.
Axis assembly occurs in rec8-6A and rec8-29A cells, but Zip1 assembly does not. (A) Examples of meiotic cells with various degrees of chromosome axis assembly as judged by Hop1 staining on chromosome spreads. Cells carry a Rec8-3HA construct. α-Hop1 is shown in green, α-HA in red, and DNA in blue. (B–E) Wild-type (A20066, ■), _rec8_Δ (A3528, ▴), rec8-29A (A14385, ·), and rec8-6A (A15042, □) cells were induced to sporulate. At the indicated times the percentage of cells with unassembled spindles (B) and chromosome spreads were assayed for Hop1 staining (C), Zip1 staining (D), and full Zip1 staining (E). The percentage of cells in the individual Zip1 categories is shown in Supplemental Figure S8. Note that cells were scored as having Hop1 assembled when they showed partial or full Hop1 staining according to the categories shown (A). n = 200 cells counted per strain per time point. Also note that in E the lines for _rec8_Δ, rec8-29A, and rec8-6A cells all overlap with each other and the _x_-axis.
Figure 8.
Effects of Cdc5 phosphorylation on Zip1 assembly and response of rec8-6A and rec8-29A cells to recombination checkpoint defects. (A, C, and E) Wild-type (A20066, ■) and cdc5-mn (A5844, ▴) cells were induced to sporulate. At the indicated times the percentage of cells with unassembled spindles (A) and chromosome spreads were assayed for Zip1 staining (C) and full Zip1 staining (E). The percentage of cells in the individual Zip1 categories is shown in Supplemental Figure S10, A and B. Note that these data are from the same experiment as is presented in Figure 7, so the wild-type control is identical in both figures. (B, D, and F) Wild-type (A14655, ■) and rec8-psa (A15364, ▴) were induced to sporulate. At the indicated times the percentage of cells with unassembled spindles (B) and chromosome spreads were assayed for Zip1 staining (D) and full Zip1 staining (F). The percentage of cells in the individual Zip1 categories is shown in Supplemental Figure S10, C and D. (G) Wild-type (A1972, ■), _mek1_Δ (A20156, ▴), rec8-6A (A15042, ·), _rec8-6A mek1_Δ (A20154, □), rec8-29A (A14385, ▵), and _rec8-29A mek1_Δ (A20157, ○) cells were induced to sporulate. At the indicated times the percentage of cells with unassembled spindles. Note that this experiment was performed concomitantly with that shown in H. Hence the controls are identical. (H) Wild-type (A1972, ■), _pch2_Δ (A21053, ▴), rec8-6A (A15042, ·), _rec8-6A pch2_Δ (A20151, □), rec8-29A (A14385, ▵), and _rec8-29A pch2_Δ (A20164, ○) cells were induced to sporulate. At the indicated times the percentage of cells with unassembled spindles.
Similar articles
- Pds5 is required for homologue pairing and inhibits synapsis of sister chromatids during yeast meiosis.
Jin H, Guacci V, Yu HG. Jin H, et al. J Cell Biol. 2009 Sep 7;186(5):713-25. doi: 10.1083/jcb.200810107. J Cell Biol. 2009. PMID: 19736318 Free PMC article. - Chromosome morphogenesis: condensin-dependent cohesin removal during meiosis.
Yu HG, Koshland D. Yu HG, et al. Cell. 2005 Nov 4;123(3):397-407. doi: 10.1016/j.cell.2005.09.014. Cell. 2005. PMID: 16269332 - Rec8 phosphorylation and recombination promote the step-wise loss of cohesins in meiosis.
Brar GA, Kiburz BM, Zhang Y, Kim JE, White F, Amon A. Brar GA, et al. Nature. 2006 May 25;441(7092):532-6. doi: 10.1038/nature04794. Epub 2006 May 3. Nature. 2006. PMID: 16672979 - Meiotic prophase-like pathway for cleavage-independent removal of cohesin for chromosome morphogenesis.
Challa K, Shinohara M, Shinohara A. Challa K, et al. Curr Genet. 2019 Aug;65(4):817-827. doi: 10.1007/s00294-019-00959-x. Epub 2019 Mar 28. Curr Genet. 2019. PMID: 30923890 Review. - Chromosome cohesion - rings, knots, orcs and fellowship.
Díaz-Martínez LA, Giménez-Abián JF, Clarke DJ. Díaz-Martínez LA, et al. J Cell Sci. 2008 Jul 1;121(Pt 13):2107-14. doi: 10.1242/jcs.029132. J Cell Sci. 2008. PMID: 18565823 Review.
Cited by
- Multiple opposing constraints govern chromosome interactions during meiosis.
Lui DY, Cahoon CK, Burgess SM. Lui DY, et al. PLoS Genet. 2013;9(1):e1003197. doi: 10.1371/journal.pgen.1003197. Epub 2013 Jan 17. PLoS Genet. 2013. PMID: 23341780 Free PMC article. - Many functions of the meiotic cohesin.
Bardhan A. Bardhan A. Chromosome Res. 2010 Dec;18(8):909-24. doi: 10.1007/s10577-010-9169-0. Epub 2010 Nov 18. Chromosome Res. 2010. PMID: 21086039 Review. - Dynamic trans interactions in yeast chromosomes.
Mirkin EV, Chang FS, Kleckner N. Mirkin EV, et al. PLoS One. 2013 Sep 30;8(9):e75895. doi: 10.1371/journal.pone.0075895. eCollection 2013. PLoS One. 2013. PMID: 24098740 Free PMC article. - Ipl1/Aurora-B is necessary for kinetochore restructuring in meiosis I in Saccharomyces cerevisiae.
Meyer RE, Chuong HH, Hild M, Hansen CL, Kinter M, Dawson DS. Meyer RE, et al. Mol Biol Cell. 2015 Sep 1;26(17):2986-3000. doi: 10.1091/mbc.E15-01-0032. Epub 2015 Jul 8. Mol Biol Cell. 2015. PMID: 26157162 Free PMC article. - Phosphorylation of chromosome core components may serve as axis marks for the status of chromosomal events during mammalian meiosis.
Fukuda T, Pratto F, Schimenti JC, Turner JM, Camerini-Otero RD, Höög C. Fukuda T, et al. PLoS Genet. 2012 Feb;8(2):e1002485. doi: 10.1371/journal.pgen.1002485. Epub 2012 Feb 9. PLoS Genet. 2012. PMID: 22346761 Free PMC article.
References
- Aguilar C., Davidson C., Dix M., Stead K., Zheng K., Hartman T., Guacci V. Topoisomerase II suppresses the temperature sensitivity of Saccharomyces cerevisiae pds5 mutants, but not the defect in sister chromatid cohesion. Cell Cycle. 2005;4:1294–1304. - PubMed
- Alexandru G., Uhlmann F., Mechtler K., Poupart M. A., Nasmyth K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell. 2001;105:459–472. - PubMed
- Bishop D. K., Park D., Xu L., Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. - PubMed
- Blitzblau H. G., Bell G. W., Rodriguez J., Bell S. P., Hochwagen A. Mapping of meiotic single-stranded DNA reveals double-stranded-break hotspots near centromeres and telomeres. Curr. Biol. 2007;17:2003–2012. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases