Chromatin-associated periodicity in genetic variation downstream of transcriptional start sites - PubMed (original) (raw)
. 2009 Jan 16;323(5912):401-4.
doi: 10.1126/science.1163183. Epub 2008 Dec 11.
Cecilia C Mello, Atsuko Shimada, Yoichiro Nakatani, Shin-Ichi Hashimoto, Masako Ogawa, Kouji Matsushima, Sam Guoping Gu, Masahiro Kasahara, Budrul Ahsan, Atsushi Sasaki, Taro Saito, Yutaka Suzuki, Sumio Sugano, Yuji Kohara, Hiroyuki Takeda, Andrew Fire, Shinichi Morishita
Affiliations
- PMID: 19074313
- PMCID: PMC2757552
- DOI: 10.1126/science.1163183
Chromatin-associated periodicity in genetic variation downstream of transcriptional start sites
Shin Sasaki et al. Science. 2009.
Abstract
Might DNA sequence variation reflect germline genetic activity and underlying chromatin structure? We investigated this question using medaka (Japanese killifish, Oryzias latipes), by comparing the genomic sequences of two strains (Hd-rR and HNI) and by mapping approximately 37.3 million nucleosome cores from Hd-rR blastulae and 11,654 representative transcription start sites from six embryonic stages. We observed a distinctive approximately 200-base pair (bp) periodic pattern of genetic variation downstream of transcription start sites; the rate of insertions and deletions longer than 1 bp peaked at positions of approximately +200, +400, and +600 bp, whereas the point mutation rate showed corresponding valleys. This approximately 200-bp periodicity was correlated with the chromatin structure, with nucleosome occupancy minimized at positions 0, +200, +400, and +600 bp. These data exemplify the potential for genetic activity (transcription) and chromatin structure to contribute to molding the DNA sequence on an evolutionary time scale.
Figures
Figure 1
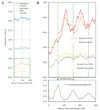
Diversity rates and nucleosome positions around TSSs. A. The x-axis shows the distance from the representative TSSs in the medaka (Hd-rR) genome. Blue line: mismatch mutation rate; light blue line: transition rate; light green line: transversion rate; red line: indel mutation rate; gray line: rate of indels of length 1bp. For smoothing of lines, a running average over a 23-bp window (one full turn of the helix in each direction) is depicted. B. The upper portion illustrates putative nucleosome dyads (red points, 73bp from start of sequence read) and cores (grey bars; 147bp). The lower table illustrates the distinct meanings of the three nucleosome indicators. C. Distribution of nucleosomes, substitutions, and indels surrounding a TSS. Black boxes: exons of the gene; blue histograms: distributions of the three nucleosome indicators; green vertical bars: substitutions between the Hd-rR and HNI genomes; red bars: deletions from the Hd-rR genome; blue bars: insertions into the Hd-rR genome; gray bars and boxes: failure of alignment. D. The green line presents the average local dyad positioning score.
Figure 2
Mutational spectra at positions around 8,181 positioned dyads that are isolated from their neighboring dyads by >165bp and are covered by an average of 5.44 putative nucleosome cores on a genome-wide scale (excluding TSSs and coding regions). A. In non-promoter regions where transcription does not occur, the two locations in the distinct strands are positionally equivalent in a nucleosome core if they are the same distance from the dyad. The x-axis presents the distance. Blue line: substitution rate; light blue line: transition rate; light green line: transversion rate; orange line: indel rate; yellow line: rate of 1bp indels. B. An expanded view of the indel rates enclosed in the green square in Fig. 2A is duplicated in tandem, and the two copies are overlaid for comparison with equivalent measurements relative to TSSs in Fig. 1A.The bottom panel presents the estimated dyads (arrows) aligned with dyad positioning score near TSSs (expanded from Fig. 1D).
Figure 3
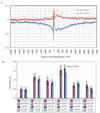
A. Base composition surrounding transcription start sites (TSSs). Red line: the difference between guanines and cytosines; blue line: the difference between adenines and thymines. B. Substitution ratio around TSSs. Rates for each substitution and its complement and their 95% confidence intervals are indicated side by side for untranscribed and transcribed regions that are upstream and downstream of TSSs, respectively.
Comment in
- Molecular biology. The structure of change.
Semple CA, Taylor MS. Semple CA, et al. Science. 2009 Jan 16;323(5912):347-8. doi: 10.1126/science.1169408. Science. 2009. PMID: 19150834 No abstract available.
Similar articles
- Molecular biology. The structure of change.
Semple CA, Taylor MS. Semple CA, et al. Science. 2009 Jan 16;323(5912):347-8. doi: 10.1126/science.1169408. Science. 2009. PMID: 19150834 No abstract available. - Associations between nucleosome phasing, sequence asymmetry, and tissue-specific expression in a set of inbred Medaka species.
Nakatani Y, Mello CC, Hashimoto S, Shimada A, Nakamura R, Tsukahara T, Qu W, Yoshimura J, Suzuki Y, Sugano S, Takeda H, Fire A, Morishita S. Nakatani Y, et al. BMC Genomics. 2015 Nov 19;16:978. doi: 10.1186/s12864-015-2198-5. BMC Genomics. 2015. PMID: 26584643 Free PMC article. - Somatic and Germline Mutation Periodicity Follow the Orientation of the DNA Minor Groove around Nucleosomes.
Pich O, Muiños F, Sabarinathan R, Reyes-Salazar I, Gonzalez-Perez A, Lopez-Bigas N. Pich O, et al. Cell. 2018 Nov 1;175(4):1074-1087.e18. doi: 10.1016/j.cell.2018.10.004. Cell. 2018. PMID: 30388444 - What positions nucleosomes?--A model.
Kiyama R, Trifonov EN. Kiyama R, et al. FEBS Lett. 2002 Jul 17;523(1-3):7-11. doi: 10.1016/s0014-5793(02)02937-x. FEBS Lett. 2002. PMID: 12123795 Review. - Histones, nucleosomes and the roles of chromatin structure in transcriptional control.
Wolffe AP. Wolffe AP. Biochem Soc Trans. 1997 May;25(2):354-8. doi: 10.1042/bst0250354. Biochem Soc Trans. 1997. PMID: 9191116 Review. No abstract available.
Cited by
- Lagging-strand replication shapes the mutational landscape of the genome.
Reijns MAM, Kemp H, Ding J, de Procé SM, Jackson AP, Taylor MS. Reijns MAM, et al. Nature. 2015 Feb 26;518(7540):502-506. doi: 10.1038/nature14183. Epub 2015 Jan 26. Nature. 2015. PMID: 25624100 Free PMC article. - Cis- and trans-regulatory divergence between progenitor species determines gene-expression novelty in Arabidopsis allopolyploids.
Shi X, Ng DW, Zhang C, Comai L, Ye W, Chen ZJ. Shi X, et al. Nat Commun. 2012 Jul 17;3:950. doi: 10.1038/ncomms1954. Nat Commun. 2012. PMID: 22805557 - NORMAL: accurate nucleosome positioning using a modified Gaussian mixture model.
Polishko A, Ponts N, Le Roch KG, Lonardi S. Polishko A, et al. Bioinformatics. 2012 Jun 15;28(12):i242-9. doi: 10.1093/bioinformatics/bts206. Bioinformatics. 2012. PMID: 22689767 Free PMC article. - Chromatin structure influences rate and spectrum of spontaneous mutations in Neurospora crassa.
Villalba de la Peña M, Summanen PAM, Liukkonen M, Kronholm I. Villalba de la Peña M, et al. Genome Res. 2023 Apr;33(4):599-611. doi: 10.1101/gr.276992.122. Epub 2023 Mar 15. Genome Res. 2023. PMID: 36922001 Free PMC article. - Impact of chromatin structure on sequence variability in the human genome.
Tolstorukov MY, Volfovsky N, Stephens RM, Park PJ. Tolstorukov MY, et al. Nat Struct Mol Biol. 2011 Apr;18(4):510-5. doi: 10.1038/nsmb.2012. Epub 2011 Mar 13. Nat Struct Mol Biol. 2011. PMID: 21399641 Free PMC article.
References
- Svejstrup J. Nat. Rev. Mol. Cell Biol. 2002;3:21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 CA09151/CA/NCI NIH HHS/United States
- R01 GM37706/GM/NIGMS NIH HHS/United States
- R01 GM037706/GM/NIGMS NIH HHS/United States
- R01 GM037706-24/GM/NIGMS NIH HHS/United States
- T32 CA009151/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials