Replication and partitioning of papillomavirus genomes - PubMed (original) (raw)
Review
Replication and partitioning of papillomavirus genomes
Alison A McBride. Adv Virus Res. 2008.
Abstract
Papillomaviruses establish persistent infection in the dividing, basal epithelial cells of the host. The viral genome is maintained as a circular, double-stranded DNA, extrachromosomal element within these cells. Viral genome amplification occurs only when the epithelial cells differentiate and viral particles are shed in squames that are sloughed from the surface of the epithelium. There are three modes of replication in the papillomavirus life cycle. Upon entry, in the establishment phase, the viral genome is amplified to a low copy number. In the second maintenance phase, the genome replicates in dividing cells at a constant copy number, in synchrony with the cellular DNA. And finally, in the vegetative or productive phase, the viral DNA is amplified to a high copy number in differentiated cells and is destined to be packaged in viral capsids. This review discusses the cis elements and protein factors required for each stage of papillomavirus replication.
Figures
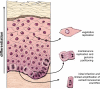
Figure 1. Modes of Replication in the Life cycle of Papillomaviruses
The diagram shows a stratified epithelium. The lowermost basal layer provides the germinal cells necessary for renewal of the epithelium. Papillomaviruses infect these cells through microabrasions and the viral DNA undergoes a limited amplification after delivery to the nucleus. The viral genome is maintained and partitioned in a regulated manner in these dividing cells. They provide a reservoir of infected cells and, as they divide, individual infected daughter cells are pushed upwards and begin the differentiation process. These differentiated cells support vegetative replication resulting in a great amplification of the viral genome. Eventually, the cells at the outermost layer, containing virion particles, are sloughed from the epithelium.
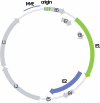
Figure 2. Papillomavirus genome
The early and late open reading frames, are indicated as E1–E8 and L1–L2, respectively. The replication elements MME (minichromosome maintenance element) and origin are shown in the LCR region. The E1 and E2 replication proteins are highlighted in green and purple, respectively, with the corresponding binding sites shown in the origin region.
Figure 3. Domain structure of the E1 and E2 proteins
A. The full-length E2 proteins of all papillomaviruses consist of two conserved domains linked by a less conserved hinge region. The N-terminal domain is important for transcriptional regulation and interaction with the E1 protein. The C-ternminal domain is a specific DNA binding and dimerization domain. Phosphorylation sites mapped in BPV-1 E2 are indicated (Lehman and Botchan, 1998; McBride et al, 1989). B. The E1 proteins consist of four domains. The N-terminal domain is important for intracellular localization and contains both nuclear localization signals (NLS) and nuclear export signals (NES). The next domain is an origin specific DNA binding domain, followed by an oligomerization domain. The C-terminal domain contains the helicase function. Phosphorylation sites mapped in BPV-1 are indicated above and those mapped in HPV11 are indicated below. See text for references.
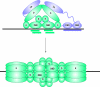
Figure 4. Initiation of DNA Replication
Initially, dimers of the E1 and E2 proteins co-operative together to bind to their specific binding sites on the replication origin. The transactivation domain of E2 interacts with the helicase domain of E1. This complex converts to a double E1 hexamer that encircles the origin. In the absence of E2, the helicase domain can make additional, non-specific interactions with DNA surrounding the origin.
Figure 5. Structures of Papillomavirus E1 and E2 Protein Domains and Complexes
A. The transactivation domain (residues 1–201) of the HPV-16 E2 protein from the pdb file 1DTO (Antson et al, 2000). Residues R37 and I73 are shown in green and residue D39 is shown in blue. B. A dimer of the DNA binding/dimerization domain of BPV-1 E2 bound to DNA (residues 326–410) from the pdb file 2BOP (Hegde et al, 1992). The DNA is shown in gray. C. The transactivation domain of HPV-18 E2 (residues 1–215; red) in complex with a c-terminal domain of HPV18 E1 (residues 428–631; green) from the pdb file 1TUE (Abbate et al, 2004). E2 residue D43 (equivalent to BPV-1 D39 in panel A) is shown in blue. D. A dimer of the BPV-1 E1 DNA binding domain (residues 159–303) from the pdb file 1FO8 (Enemark et al, 2000). Residues shown in blue are important for DNA contact. E. Double hexameric ring structure of the BPV-1 E1 helicase domain (residues 306–577) from the pdb file 2GXA (Enemark and Joshua-Tor, 2006). Each E1 monomer is shown in a different color.
Figure 6. E1 Binding Origin Sequences
The palindromic E1 binding origin sequence from a range of papillomaviruses is shown. The genus to which each virus belongs is shown to the right.
Figure 7. Map of the E1 and E2 binding sites in the Long Control Regions in a Range of Papillomaviruses
The LCRs of papillomaviruses from five different genera are shown. The end of the L1 and beginning of the E6 open reading frames are indicated. The E1 binding site is indicated as a green box. E2 binding sites that match the consensus ACCN6GGT are shown in purple. E2 sites that deviate from this consensus are shown in hatched purple.
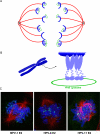
Figure 8. Mechanism of Papillomavirus Genome Partitioning
A. Model for partitioning of papillomavirus genomes. Cellular chromosomes (blue crescents) are partitioned by attachment to the spindle in mitotis. The viral E2 protein (purple) binds to the viral genome and tethers it to mitotic chromosomes, thus hitchhiking on the cellular chromosomes. B. Model for chromosomal tethering by the BPV-1 E2 protein. The E2 DNA binding domain binds to sites in the viral genome while the transactivation domain interacts with proteins, such as Brd4, on mitotic chromosomes. C. Binding patterns of three different E2 proteins, as detected by immunofluorescence, on mitotic chromosomes. BPV-1 E2 forms small speckles on the arms of all chromosomes. HPV-8 E2 is observed in fewer, larger speckles that bind adjacent to the centromere of acrocentric chromosomes. HPV-11 E2 is also observed binding to the pericentromeric region of chromosomes, but only in certain fixation conditions (Oliveira et al, 2006). The E2 proteins are shown in green, cellular DNA in blue and the mitotic spindle in red.
Figure 9. Vegetative DNA amplification in a BPV-1 Papilloma
E2-specific immunofluorescence and BPV DNA-specific fluorescent in situ hybridization were performed on serial sections of bovine wart tissue. E2 protein expression is shown in green in the top panels and viral DNA is shown in green in the bottom panels. In both cases cellular DNA is counterstained in red. Cells containing high levels of both E2 protein and viral DNA are indicate by arrows.
Similar articles
- Persistence of an Oncogenic Papillomavirus Genome Requires cis Elements from the Viral Transcriptional Enhancer.
Van Doorslaer K, Chen D, Chapman S, Khan J, McBride AA. Van Doorslaer K, et al. mBio. 2017 Nov 21;8(6):e01758-17. doi: 10.1128/mBio.01758-17. mBio. 2017. PMID: 29162712 Free PMC article. - Persistent Human Papillomavirus Infection.
Della Fera AN, Warburton A, Coursey TL, Khurana S, McBride AA. Della Fera AN, et al. Viruses. 2021 Feb 20;13(2):321. doi: 10.3390/v13020321. Viruses. 2021. PMID: 33672465 Free PMC article. Review. - Mechanisms and strategies of papillomavirus replication.
McBride AA. McBride AA. Biol Chem. 2017 Jul 26;398(8):919-927. doi: 10.1515/hsz-2017-0113. Biol Chem. 2017. PMID: 28315855 Review. - Papillomavirus DNA replication - from initiation to genomic instability.
Kadaja M, Silla T, Ustav E, Ustav M. Kadaja M, et al. Virology. 2009 Feb 20;384(2):360-8. doi: 10.1016/j.virol.2008.11.032. Epub 2009 Jan 13. Virology. 2009. PMID: 19141359 Review. - Human Papillomavirus Genome Copy Number Is Maintained by S-Phase Amplification, Genome Loss to the Cytosol during Mitosis, and Degradation in G1 Phase.
Bienkowska-Haba M, Zwolinska K, Keiffer T, Scott RS, Sapp M. Bienkowska-Haba M, et al. J Virol. 2023 Feb 28;97(2):e0187922. doi: 10.1128/jvi.01879-22. Epub 2023 Feb 7. J Virol. 2023. PMID: 36749071 Free PMC article.
Cited by
- A Flp-SUMO hybrid recombinase reveals multi-layered copy number control of a selfish DNA element through post-translational modification.
Ma CH, Su BY, Maciaszek A, Fan HF, Guga P, Jayaram M. Ma CH, et al. PLoS Genet. 2019 Jun 26;15(6):e1008193. doi: 10.1371/journal.pgen.1008193. eCollection 2019 Jun. PLoS Genet. 2019. PMID: 31242181 Free PMC article. - Replication interference between human papillomavirus types 16 and 18 mediated by heterologous E1 helicases.
Mori S, Kusumoto-Matsuo R, Ishii Y, Takeuchi T, Kukimoto I. Mori S, et al. Virol J. 2014 Jan 24;11:11. doi: 10.1186/1743-422X-11-11. Virol J. 2014. PMID: 24456830 Free PMC article. - Epidermal Growth Factor Receptor and Abl2 Kinase Regulate Distinct Steps of Human Papillomavirus 16 Endocytosis.
Bannach C, Brinkert P, Kühling L, Greune L, Schmidt MA, Schelhaas M. Bannach C, et al. J Virol. 2020 May 18;94(11):e02143-19. doi: 10.1128/JVI.02143-19. Print 2020 May 18. J Virol. 2020. PMID: 32188731 Free PMC article. - Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability.
Akagi K, Li J, Broutian TR, Padilla-Nash H, Xiao W, Jiang B, Rocco JW, Teknos TN, Kumar B, Wangsa D, He D, Ried T, Symer DE, Gillison ML. Akagi K, et al. Genome Res. 2014 Feb;24(2):185-99. doi: 10.1101/gr.164806.113. Epub 2013 Nov 7. Genome Res. 2014. PMID: 24201445 Free PMC article. - Assessing parallel gene histories in viral genomes.
Mengual-Chuliá B, Bedhomme S, Lafforgue G, Elena SF, Bravo IG. Mengual-Chuliá B, et al. BMC Evol Biol. 2016 Feb 5;16:32. doi: 10.1186/s12862-016-0605-4. BMC Evol Biol. 2016. PMID: 26847371 Free PMC article.
References
- Abbate EA, Voitenleitner C, Botchan MR. Structure of the papillomavirus DNA-tethering complex E2:Brd4 and a peptide that ablates HPV chromosomal association. Mol.Cell. 2006;24:877–889. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- Z01 AI000713/ImNIH/Intramural NIH HHS/United States
- ZIA AI001073/ImNIH/Intramural NIH HHS/United States
- ZIA AI000713/ImNIH/Intramural NIH HHS/United States
- Z01 AI001072/ImNIH/Intramural NIH HHS/United States
- ZIA AI001072/ImNIH/Intramural NIH HHS/United States
- Z99 AI999999/ImNIH/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources