Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions - PubMed (original) (raw)
Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions
Sherwood R Casjens et al. Methods Mol Biol. 2009.
Abstract
Tailed-bacteriophage virions contain a single linear dsDNA chromosome which can range in size from about 18 to 500 kbp across the known tailed-phage types. These linear chromosomes can have one of several known types of termini as follows: cohesive ends (5'- or 3'-single-strand extensions), circularly permuted direct terminal repeats, short or long exact direct terminal repeats, terminal host DNA sequences, or covalently bound terminal proteins. These different types of ends reflect differing DNA replication strategies and especially differing terminase actions during DNA packaging. In general, complete genome sequence determination does not by itself elucidate the nature of these ends, so directed experimental analysis is usually required to understand the nature of the virion chromosome ends. This chapter discusses these methods.
Figures
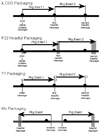
Fig. 7.1
Four tailed-phage DNA packaging strategies. Packaging strategies of phages λ, P22, T7, and Mu are shown diagrammatically. Thick black horizontal lines represent phage concatemeric DNAs, or, in the case of Mu, phage genomes that are integrated into the host bacteria’s chromosome (the latter represented by thick gray line). Black circles mark the packaging recognition sites and horizontal black arrows represent individual packaging events. Vertical black lines indicate precise terminase cleavages and vertical gray lines indicate imprecise cleavages (see text). In each case except Mu, sequential series of packaging events occur, in which subsequent events (event 2 in figure) on the same concatemer molecule begin at the concatemer end created by the previous event (event 1); although only two successive events are shown, packaging series can in some cases be up to 10 or more events long. In phages λ and T7, each event begins and ends at a packaging recognition site, and in phage T7 the white rectangles show the region (the direct terminal repeat) that is duplicated in concert with packaging. In phage P22, the increasing width of the vertical gray boxes to the right, denotes the increased range of cleavage site locations as events proceed rightward. The small gray horizontal rectangle below the first P22 event is the optimal location of the Southern probe used to analyze pac fragments (see text).
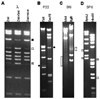
Fig. 7.2
Restriction enzyme generated fragments from tailed-phage virion DNA. DNA fragments were separated by 0.8% agarose gel electrophoresis and visualized by staining with ethidium bromide. The phage virion source of the DNA is indicated above each panel, and the restriction enzymes used are indicated above each lane. A. Phage λ DNA after normal isolation and storage. DNA in the second and third lanes was heated to 75 °C for 15 min and then fast or slow cooled to room temperature, respectively, as described in the Methods. Terminal DNA fragments are indicated as follows: white square, left end fragment; gray square, right end fragment; black square, left and right end fragments annealed together by their cohesive ends. B. Phage P22 DNA. The pac fragments generated by PstI and EcoRI are indicated by black circles on the left and right, respectively. C. Phage Sf6 DNA. The locations of the diffuse pac fragment bands generated by imprecise packaging series initiation are indicated by white rectangles (see also ref. (31)). D. Phage SF6 DNA. Terminal DNA fragments are indicated as follows: white square, left end fragment; gray square, right end fragment.
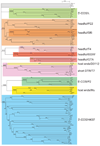
Fig. 7.3
Neighbor-joining tree of large terminase subunit amino acid sequences. A neighbor-joining tree of 123 tailed-phage terminase amino acid sequences was generated by CLUSTAL X (59). The numbers near bifurcations are bootstrap values for 1,000 trials. Short bifurcating branches linking the major groups shown here were manually merged, since all had low bootstrap values, and the major deep branches are all shown as radiating from a single source. The names of the phages or prophages are shown at the right of each terminal branch (some prophage names are those proposed by Casjens (13)). Major robust, related groups of terminases are highlighted with gray boxes, and the packaging strategy and a prototype phage for each group is given at the far right. Those phages whose virion DNA termini structures have been experimentally determined are indicated as follows: #, 5′-cohesive ends in lambdoid phages;^, 5′-cohesive ends in P2-like phages; *, 3′-cohesive ends; $, T7-like phages with direct terminal repeats and no circular permutation; , phages with long direct terminal repeats and no circular permutation; @, phages with host DNA at termini; †, headful packaging in non-T4-like phages, including P22 and the gene transfer agents (GTA); %, headful packaging in T4-like phages; ●, headful packaging phages for which no obvious pac fragment band has been identified in ethidium stained electrophoresis gels of restricted DNA (see text; the darker orange boxed “headful/Sf6” subgroup within the “headful/P22” group appear to be one branch of this type of terminase). This figure is modified from Fig. 7.6 of Casjens et al. (32).
Similar articles
- DNA Packaging and Genomics of the Salmonella 9NA-Like Phages.
Zeng C, Gilcrease EB, Hendrix RW, Xie Y, Jalfon MJ, Gill JJ, Casjens SR. Zeng C, et al. J Virol. 2019 Oct 29;93(22):e00848-19. doi: 10.1128/JVI.00848-19. Print 2019 Nov 15. J Virol. 2019. PMID: 31462565 Free PMC article. - Software-based analysis of bacteriophage genomes, physical ends, and packaging strategies.
Merrill BD, Ward AT, Grose JH, Hope S. Merrill BD, et al. BMC Genomics. 2016 Aug 26;17(1):679. doi: 10.1186/s12864-016-3018-2. BMC Genomics. 2016. PMID: 27561606 Free PMC article. - DNA Topology and the Initiation of Virus DNA Packaging.
Oh CS, Sippy J, Charbonneau B, Crow Hutchinson J, Mejia-Romero OE, Barton M, Patel P, Sippy R, Feiss M. Oh CS, et al. PLoS One. 2016 May 4;11(5):e0154785. doi: 10.1371/journal.pone.0154785. eCollection 2016. PLoS One. 2016. PMID: 27144448 Free PMC article. - Virus DNA packaging: the strategy used by phage lambda.
Catalano CE, Cue D, Feiss M. Catalano CE, et al. Mol Microbiol. 1995 Jun;16(6):1075-86. doi: 10.1111/j.1365-2958.1995.tb02333.x. Mol Microbiol. 1995. PMID: 8577244 Review. - The dsDNA packaging motor in bacteriophage ø29.
Morais MC. Morais MC. Adv Exp Med Biol. 2012;726:511-47. doi: 10.1007/978-1-4614-0980-9_23. Adv Exp Med Biol. 2012. PMID: 22297529 Review.
Cited by
- Tyroviruses are a new group of temperate phages that infect Bacillus species in soil environments worldwide.
Batinovic S, Stanton CR, Rice DTF, Rowe B, Beer M, Petrovski S. Batinovic S, et al. BMC Genomics. 2022 Nov 28;23(1):777. doi: 10.1186/s12864-022-09023-4. BMC Genomics. 2022. PMID: 36443683 Free PMC article. - A novel virulent Litunavirus phage possesses therapeutic value against multidrug resistant Pseudomonas aeruginosa.
Lerdsittikul V, Thongdee M, Chaiwattanarungruengpaisan S, Atithep T, Apiratwarrasakul S, Withatanung P, Clokie MRJ, Korbsrisate S. Lerdsittikul V, et al. Sci Rep. 2022 Dec 7;12(1):21193. doi: 10.1038/s41598-022-25576-6. Sci Rep. 2022. PMID: 36476652 Free PMC article. - Complete genome sequence of enterococcal phage G01.
Sheriff EK, Andersen SE, Chatterjee A, Duerkop BA. Sheriff EK, et al. Microbiol Resour Announc. 2024 Mar 12;13(3):e0121723. doi: 10.1128/mra.01217-23. Epub 2024 Jan 31. Microbiol Resour Announc. 2024. PMID: 38294211 Free PMC article. - Virus genomics: what is being overlooked?
Kieft K, Anantharaman K. Kieft K, et al. Curr Opin Virol. 2022 Apr;53:101200. doi: 10.1016/j.coviro.2022.101200. Epub 2022 Jan 17. Curr Opin Virol. 2022. PMID: 35051682 Free PMC article. Review. - Listeria phages: Genomes, evolution, and application.
Klumpp J, Loessner MJ. Klumpp J, et al. Bacteriophage. 2013 Jul 1;3(3):e26861. doi: 10.4161/bact.26861. Epub 2013 Oct 24. Bacteriophage. 2013. PMID: 24251077 Free PMC article. Review.
References
- Mousset S, Thomas R. Ter, a function which generates the ends of the mature lambda chromosome. Nature. 1969;221:242–244. - PubMed
- Feiss M, Campbell A. Duplication of the bacteriophage lambda cohesive end site: genetic studies. J. Mol. Biol. 1974;83:527–540. - PubMed
- Jackson EN, Jackson DA, Deans RJ. EcoRI analysis of bacteriophage P22 DNA packaging. J. Mol. Biol. 1978;118:365–388. - PubMed
- Emmons SW. Bacteriophage lambda derivatives carrying two copies of the cohesive end site. J. Mol. Biol. 1974;83:511–525. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources