miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels - PubMed (original) (raw)
miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels
Kotb Abdelmohsen et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Gene expression is potently regulated through the action of RNA-binding proteins (RBPs) and microRNAs (miRNAs). Here, we present evidence of a miRNA regulating an RBP. The RBP HuR can stabilize and modulate the translation of numerous target mRNAs involved in cell proliferation, but little is known about the mechanisms that regulate HuR abundance. We identified two putative sites of miR-519 interaction on the HuR mRNA, one in its coding region (CR), one in its 3'-untranslated region (UTR). In several human carcinoma cell lines tested, HeLa (cervical), HCT116 and RKO (colon), and A2780 (ovarian), overexpression of a miR-519 precursor [(Pre)miR-519] reduced HuR abundance, while inhibiting miR-519 by using an antisense RNA [(AS)miR-519] elevated HuR levels. The influence of miR-519 was recapitulated using heterologous reporter constructs that revealed a greater repressive effect on the HuR CR than the HuR 3'-UTR target sequences. miR-519 did not alter HuR mRNA abundance, but reduced HuR biosynthesis, as determined by measuring nascent HuR translation and HuR mRNA association with polysomes. Modulation of miR-519 leading to altered HuR levels in turn affected the levels of proteins encoded by HuR target mRNAs. In keeping with HuR's proliferative influence, (AS)miR-519 significantly increased cell number and [(3)H]-thymidine incorporation, while (Pre)miR-519 reduced these parameters. Importantly, the growth-promoting effects of (AS)miR-519 required the presence of HuR, because downregulation of HuR by RNAi dramatically suppressed its proliferative action. In sum, miR-519 represses HuR translation, in turn reducing HuR-regulated gene expression and cell division.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
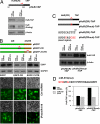
miR-519 influences HuR levels. (A) Schematic of HuR mRNA depicting two predicted target sites for the miR-519 family within the CR and 3′-UTR. Alignment of the HuR mRNA sequences with three members of the miR-519 family; top strand, HuR mRNA; bottom strand, miR-519. (B) The effect of three miR-519 family members on HuR levels was tested by transfecting HeLa cells with the corresponding precursor (Pre) and antisense (AS) transcripts, and with control miRNAs. By 48 h after transfection, the levels of HuR and loading control β-actin were tested in HeLa whole-cell lysates by Western blotting. (C) miR-519 (miR-519b-3p) abundance as measured by RT-qPCR 48 h after transfection of HeLa cells with Ctrl miRNA, (Pre)miR-519, or (AS)miR-519; 18S rRNA levels were used to monitor loading differences. Graph shows representative results from two experiments. (D) In the cell lines shown, HuR and β-actin levels were analyzed by Western blotting (Top) and miR-519 levels (Bottom) were measured as described in C. Graph shows the means and standard deviation (SD) from three independent experiments. (E) The modulation of miR-519 in the colon carcinoma lines RKO and HCT116, and the analysis of HuR and β-actin were performed as described in B. (F) The levels of HuR and loading control β-actin were examined by Western blotting in the ovarian epithelial line HOSE-B and the ovarian carcinoma line A2780. (G) The levels of HuR mRNA and miR-519 (each normalized to 18S rRNA levels), were analyzed by RT-qPCR. (H) The effect of modulating miR-519 levels in HOSE-B and A2780 cells was studied as described in E.
Fig. 2.
Influence of miR-519 on HuR expression. HeLa cells were transfected with the indicated siRNAs or miRNAs; 48 h later, the levels of HuR and loading control β-actin were assessed by Western blotting (A) and the levels of HuR mRNA and normalization control 18S rRNA were quantified by RT-qPCR (B). (C) De novo HuR biosynthesis, as assessed by HuR IP 48 h after transfection of the indicated small RNAs (details in Materials and Methods). (D) Cells that were transfected as described in C were fractionated through sucrose gradients and the relative distribution of HuR mRNA (and housekeeping GAPDH mRNA) was studied by RT-qPCR analysis of RNA in each of 10 gradient fractions; 40S and 60S, small and large ribosome subunits, respectively; 80S, monosome; LMW and HMW, low- and high-molecular weight polysomes, respectively. Data are representative of three independent experiments. (E) HeLa cells were cotransfected with plasmids (expressing either HA or HA-Ago1) and with either Ctrl miRNA or (Pre)miR-519; 48 h later, the association of HuR mRNA with HA-Ago1 was assessed in IP samples prepared using anti-HA antibody. The levels of housekeeping UBC mRNA served to normalize sample input.
Fig. 3.
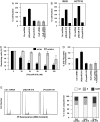
miR-519 influence on HuR reporter constructs. (A) Plasmids pHuR-TAP (expressing the HuR CR linked to a TAP tag) or control pTAP were cotransfected with Ctrl miRNA or (Pre)miR-519; the levels of endogenous HuR (Endog.) and the ectopic protein HuR-TAP were then analyzed by Western blotting. (B) Schematic of EGFP reporter constructs bearing either no HuR mRNA sequences (pEGFP), the miR-519 target site on the HuR CR (pEGFP-CR), or the miR-519 target site on the HuR 3′-UTR (pEGFP-3′-UTR); 48 h after cotransfection of the plasmids with Ctrl miRNA or (Pre)miR-519, the levels of EGFP expressed in each transfection group were analyzed by Western blotting and fluorescence microscopy. Data are representative of three independent experiments. (C Upper) Schematic of the predicted miR-519 binding site on the HuR CR; [pHuR(CR)-TAP], plasmid with the wild-type sequence; [pHuR(CRmut)-TAP], plasmid with four mutations in the miR-519 seed region. (Lower), Western blot analysis of GFP expression 48 h after cotransfection of pHuR(CR)-TAP or pHuR(CRmut)-TAP along with either Ctrl miRNA or (Pre)miR-519. (D) Top, miR-519(mut) depicting compensatory base changes (red) to restore binding to the mutated HuR(CR)-TAP mRNA. Plasmids pHuR(CR)-TAP and pHuR(CRmut)-TAP were cotransfected with either Ctrl miRNA or miR-519(mut); graph, quantification of representative HuR-TAP signals (
Fig. S5
).
Fig. 4.
Influence of miR-519-regulated HuR on cell proliferation. (A) By 48 h after transfection of HeLa cells with the siRNAs and miRNAs shown, cell numbers were measured using a hemocytometer and represented as percentage of cells relative to the Ctrl miRNA group. (B) RKO and HCT116 cells were transfected as indicated; cell numbers were quantified 48 h later as explained in A. (C) Influence of (Pre)miR-519 on the viability of ovarian cancer cells (A2780) and ovarian epithelial cells (HOSE-B) 48 h after transfection with increasing doses of (Pre)miR-519, as quantified using the MTT assay. (D) Measurement of [3H]-thymidine incorporation by 48 h after transfection of HeLa cells with the siRNAs and miRNAs indicated. The data in panels A, C, and D are the means ± SD from three experiments. (E) Forty-eight h after transfection with Ctrl miRNA, (AS)miR-519, or (Pre)miR-519, HeLa cells were subjected to FACS analysis (Left) and the relative G1, S, and G2/M compartments calculated (Right). Data are representative of three independent experiments.
Similar articles
- Competitive regulation of nucleolin expression by HuR and miR-494.
Tominaga K, Srikantan S, Lee EK, Subaran SS, Martindale JL, Abdelmohsen K, Gorospe M. Tominaga K, et al. Mol Cell Biol. 2011 Oct;31(20):4219-31. doi: 10.1128/MCB.05955-11. Epub 2011 Aug 22. Mol Cell Biol. 2011. PMID: 21859890 Free PMC article. - Post-transcriptional regulation of Wnt co-receptor LRP6 and RNA-binding protein HuR by miR-29b in intestinal epithelial cells.
Li Y, Chen G, Wang JY, Zou T, Liu L, Xiao L, Chung HK, Rao JN, Wang JY. Li Y, et al. Biochem J. 2016 Jun 1;473(11):1641-9. doi: 10.1042/BCJ20160057. Epub 2016 Apr 18. Biochem J. 2016. PMID: 27089893 Free PMC article. - Loss of repression of HuR translation by miR-16 may be responsible for the elevation of HuR in human breast carcinoma.
Xu F, Zhang X, Lei Y, Liu X, Liu Z, Tong T, Wang W. Xu F, et al. J Cell Biochem. 2010 Oct 15;111(3):727-34. doi: 10.1002/jcb.22762. J Cell Biochem. 2010. PMID: 20626035 - Properties of the Regulatory RNA-Binding Protein HuR and its Role in Controlling miRNA Repression.
Meisner NC, Filipowicz W. Meisner NC, et al. Adv Exp Med Biol. 2011;700:106-23. doi: 10.1007/978-1-4419-7823-3_10. Adv Exp Med Biol. 2011. PMID: 21755477 Review. - Properties of the regulatory RNA-binding protein HuR and its role in controlling miRNA repression.
Meisner NC, Filipowicz W. Meisner NC, et al. Adv Exp Med Biol. 2010;700:106-23. Adv Exp Med Biol. 2010. PMID: 21627034 Review.
Cited by
- HIF-1α overexpression in ductal carcinoma in situ of the breast in BRCA1 and BRCA2 mutation carriers.
van der Groep P, van Diest PJ, Smolders YH, Ausems MG, van der Luijt RB, Menko FH, Bart J, de Vries EG, van der Wall E. van der Groep P, et al. PLoS One. 2013;8(2):e56055. doi: 10.1371/journal.pone.0056055. Epub 2013 Feb 8. PLoS One. 2013. PMID: 23409121 Free PMC article. - miRNA expressions in rectal cancer as predictors of response to neoadjuvant chemoradiation therapy.
Kheirelseid EA, Miller N, Chang KH, Curran C, Hennessey E, Sheehan M, Newell J, Lemetre C, Balls G, Kerin MJ. Kheirelseid EA, et al. Int J Colorectal Dis. 2013 Feb;28(2):247-60. doi: 10.1007/s00384-012-1549-9. Epub 2012 Aug 18. Int J Colorectal Dis. 2013. PMID: 22903298 - miR-195 competes with HuR to modulate stim1 mRNA stability and regulate cell migration.
Zhuang R, Rao JN, Zou T, Liu L, Xiao L, Cao S, Hansraj NZ, Gorospe M, Wang JY. Zhuang R, et al. Nucleic Acids Res. 2013 Sep;41(16):7905-19. doi: 10.1093/nar/gkt565. Epub 2013 Jun 26. Nucleic Acids Res. 2013. PMID: 23804758 Free PMC article. - Modulation of miRNA function by natural and synthetic RNA-binding proteins in cancer.
Vos PD, Leedman PJ, Filipovska A, Rackham O. Vos PD, et al. Cell Mol Life Sci. 2019 Oct;76(19):3745-3752. doi: 10.1007/s00018-019-03163-9. Epub 2019 Jun 4. Cell Mol Life Sci. 2019. PMID: 31165201 Free PMC article. Review. - MicroRNAs: Meta-controllers of gene expression in synaptic activity emerge as genetic and diagnostic markers of human disease.
Ceman S, Saugstad J. Ceman S, et al. Pharmacol Ther. 2011 Apr;130(1):26-37. doi: 10.1016/j.pharmthera.2011.01.004. Epub 2011 Jan 20. Pharmacol Ther. 2011. PMID: 21256154 Free PMC article. Review.
References
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. - PubMed
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. - PubMed
- Abdelmohsen K, Lal A, Kim HH, Gorospe M. Posttranscriptional orchestration of an anti-apoptotic program by HuR. Cell Cycle. 2007;6:1288–1292. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous