Multilayer three-dimensional super resolution imaging of thick biological samples - PubMed (original) (raw)
Multilayer three-dimensional super resolution imaging of thick biological samples
Alipasha Vaziri et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Recent advances in optical microscopy have enabled biological imaging beyond the diffraction limit at nanometer resolution. A general feature of most of the techniques based on photoactivated localization microscopy (PALM) or stochastic optical reconstruction microscopy (STORM) has been the use of thin biological samples in combination with total internal reflection, thus limiting the imaging depth to a fraction of an optical wavelength. However, to study whole cells or organelles that are typically up to 15 microm deep into the cell, the extension of these methods to a three-dimensional (3D) super resolution technique is required. Here, we report an advance in optical microscopy that enables imaging of protein distributions in cells with a lateral localization precision better than 50 nm at multiple imaging planes deep in biological samples. The approach is based on combining the lateral super resolution provided by PALM with two-photon temporal focusing that provides optical sectioning. We have generated super-resolution images over an axial range of approximately 10 microm in both mitochondrially labeled fixed cells, and in the membranes of living S2 Drosophila cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
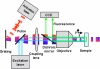
Experimental setup for temporal focusing PALM. The activation light is provided by a two-photon temporally focused beam and the excitation light by a one-photon laser. Temporal focusing of the propagating pulse is achieved by imaging a spot on a grating onto the specimen via a two lens system (objective plus coupling lens). This 4f imaging configuration ensures that the pulse has the same (shortest) width in the specimen as at the grating, but the spatial and temporal dispersion caused by the grating generates a longer pulse outside the focal region.
Fig. 2.
Visualization of dendritic spines using temporal focusing. The depth of focus for a temporally focused beam with a spot size of ≈ 15 μm is shown. It is ≈ 1.9 μm, which is ≈ 75 times smaller than for the same spot size without temporal focusing (see
SI
Materials and Methods). Individual dendritic spines at different locations of a branched dendrite are visualized using temporal focusing. Top left image shows the membrane boundary of the two dendrites whereas the rest of images show the individual spines at I, II, and III coming in and going out of focus (see
Movie S1
for the animation >100 μm). Images were taken at 10-μm steps in an acute hippocampal slice. Because the spines contained a lower concentration of dye compared with dendrites, the laser intensity was increased for the visualization of individual spines, which resulted in the saturation of the image at the dendrites.
Fig. 3.
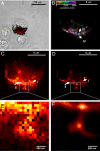
Multilayer super resolution imaging of mitochondrial network >5 μm in depth. (A) Overview of the region where the images were taken at 4× lower magnification than (B–G). The DIC images of the cell are overlaid with an epifluorescent image taken by a Xenon lamp and subsequently overlaid with (D) (red region). (B–F) PALM images of mitochondrial networks in HFF1-cell labeled with Dronpa taken in 1-μm steps. Different axial regions of the same mitochondria (region marked between I and II in B–E) and individual mitochondria (III in D) can be seen at different focal depths. (D and H) Comparison of the PALM image and the sum of diffraction-limited images for the same layer. (J and I) Five-fold magnification of a selected region from D and H, respectively. Several features seen as individual structures in J (IV and V) are clearly below the resolution limit in at the respective locations in I. (G) Superposition of (B–F) where the axial position is represented using a color map. (G) The 3D topology of the mitochondrial network. Interestingly, there are several features which are laterally as close as ≈ 200 nm but originate from planes that are axially 3 μm apart (structures within the circle in G) The volume information in (G) was further used to create a 3D animation (See
SI Movie S2
).
Fig. 4.
Multilayer PALM of Drosophila S2 cells. (A) Stack of PALM images at six axial layers separated by 1 μm each are taken from membrane labeled Drosophila S2 cells. The axial position of each layer is used for color coding and the superimposed images are shown in (B). The color information clearly shows the axial structure of the cell membrane showing a sharp axial edge between III and IV of ≈ 2.5 μm over a lateral extension of less than ≈ 200 nm. (C and D) Comparison between PALM and diffraction-limited images for one of the layers. Although the area marked as I is resolved in the PALM image (D), it is clearly below the diffraction limit in (C). The area II in (D) shows a ring type structure that is not resolved in (C). Close-ups (E and F) show protrusions from the membrane. These are individually resolved in (F) and are below the diffraction limit in (E). (A) is a DIC/PALM overlay, showing the S2 cell and the region PALM-imaged in (B–F).
Similar articles
- Recent advances in super-resolution fluorescence imaging and its applications in biology.
Han R, Li Z, Fan Y, Jiang Y. Han R, et al. J Genet Genomics. 2013 Dec 20;40(12):583-95. doi: 10.1016/j.jgg.2013.11.003. Epub 2013 Nov 23. J Genet Genomics. 2013. PMID: 24377865 Review. - Super-resolution imaging by localization microscopy.
Owen DM, Magenau A, Williamson DJ, Gaus K. Owen DM, et al. Methods Mol Biol. 2013;950:81-93. doi: 10.1007/978-1-62703-137-0_6. Methods Mol Biol. 2013. PMID: 23086871 - 3D super-resolution imaging by localization microscopy.
Magenau A, Gaus K. Magenau A, et al. Methods Mol Biol. 2015;1232:123-36. doi: 10.1007/978-1-4939-1752-5_11. Methods Mol Biol. 2015. PMID: 25331133 - Multicolor 3D super-resolution imaging by quantum dot stochastic optical reconstruction microscopy.
Xu J, Tehrani KF, Kner P. Xu J, et al. ACS Nano. 2015 Mar 24;9(3):2917-25. doi: 10.1021/nn506952g. Epub 2015 Feb 27. ACS Nano. 2015. PMID: 25703291 - A new wave of cellular imaging.
Toomre D, Bewersdorf J. Toomre D, et al. Annu Rev Cell Dev Biol. 2010;26:285-314. doi: 10.1146/annurev-cellbio-100109-104048. Annu Rev Cell Dev Biol. 2010. PMID: 20929313 Review.
Cited by
- Lightsheet localization microscopy enables fast, large-scale, and three-dimensional super-resolution imaging.
Lu CH, Tang WC, Liu YT, Chang SW, Wu FCM, Chen CY, Tsai YC, Yang SM, Kuo CW, Okada Y, Hwu YK, Chen P, Chen BC. Lu CH, et al. Commun Biol. 2019 May 9;2:177. doi: 10.1038/s42003-019-0403-9. eCollection 2019. Commun Biol. 2019. PMID: 31098410 Free PMC article. - Fluorescence nanoscopy. Methods and applications.
Requejo-Isidro J. Requejo-Isidro J. J Chem Biol. 2013 Jun 4;6(3):97-120. doi: 10.1007/s12154-013-0096-3. eCollection 2013 Jun 4. J Chem Biol. 2013. PMID: 24432127 Free PMC article. Review. - Wide-field multiphoton imaging of cellular dynamics in thick tissue by temporal focusing and patterned illumination.
Therrien OD, Aubé B, Pagès S, Koninck PD, Côté D. Therrien OD, et al. Biomed Opt Express. 2011 Feb 25;2(3):696-704. doi: 10.1364/BOE.2.000696. Biomed Opt Express. 2011. PMID: 21412473 Free PMC article. - Approaches toward super-resolution fluorescence imaging of mitochondrial proteins using PALM.
Brown TA, Fetter RD, Tkachuk AN, Clayton DA. Brown TA, et al. Methods. 2010 Aug;51(4):458-63. doi: 10.1016/j.ymeth.2010.01.001. Epub 2010 Jan 12. Methods. 2010. PMID: 20060907 Free PMC article. Review. - Light-sheet confined super-resolution using two-photon photoactivation.
Cella Zanacchi F, Lavagnino Z, Faretta M, Furia L, Diaspro A. Cella Zanacchi F, et al. PLoS One. 2013 Jul 2;8(7):e67667. doi: 10.1371/journal.pone.0067667. Print 2013. PLoS One. 2013. PMID: 23844052 Free PMC article.
References
- Juette MF, et al. Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nat Methods. 2008;5:527–529. - PubMed
- Schmidt R, et al. Spherical nanosized focal spot unravels the interior of cells. Nat Methods. 2008;5:539–544. - PubMed
- Gustafsson MGL. Super-resolution light microscopy goes live. Nat Methods. 2008;5:385–387. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases