Analysis of nondegradative protein ubiquitylation with a monoclonal antibody specific for lysine-63-linked polyubiquitin - PubMed (original) (raw)
. 2008 Dec 23;105(51):20197-202.
doi: 10.1073/pnas.0810461105. Epub 2008 Dec 17.
Atsushi Matsuzawa, Scott A Brown, JingRan Zhou, Cliff S Guy, Ping-Hui Tseng, Karen Forbes, Thomas P Nicholson, Paul W Sheppard, Hans Häcker, Michael Karin, Dario A A Vignali
Affiliations
- PMID: 19091944
- PMCID: PMC2629300
- DOI: 10.1073/pnas.0810461105
Analysis of nondegradative protein ubiquitylation with a monoclonal antibody specific for lysine-63-linked polyubiquitin
Haopeng Wang et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Modification of proteins by the addition of lysine (K)-63-linked polyubiquitin (polyUb) chains is suggested to play important roles in a variety of cellular events, including DNA repair, signal transduction, and receptor endocytosis. However, identifying such modifications in living cells is complex and cumbersome. We have generated a monoclonal antibody (mAb) that specifically recognizes K63-linked polyUb, but not any other isopeptide-linked (K6, K11, K27, K29, K33, or K48) polyUb or monoubiquitin. We demonstrate the sensitivity and specificity of this K63Ub-specific mAb to detect K63Ub-modified proteins in cell lysates by Western blotting and in cells by immunofluorescence, and K63Ub-modified TRAF6 and MEKK1 in vitro and ex vivo. This unique mAb will facilitate the analysis of K63-linked polyubiquitylation ex vivo and presents a strategy for the generation of similar reagents against other forms of polyUb.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
PolyUb structure and immunogen design. (A–C) Structure of a single Ub. (A) Protein Data Bank (PDB) coordinates: N-terminal Ub in 2JF5. (B) K63-linked di-Ub (PDB ID code 2JF5). (C) K48-linked di-Ub (PDB ID code 1AAR). Light green, N-terminal Ub; dark green, C-terminal Ub; red, K63; pink, K48. Rendering was performed by using PyMol. (D) Sequence of Ub with K63 colored red and remaining 6 lysine residues colored blue. (E and F) Diagram representations of K63-linked (E) and K48-linked di-Ub (F). Coloring is as in A–C. The residues contained within the U63U immunogen are depicted in red, whereas the corresponding residues in the U48U control branched peptide are in pink.
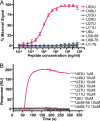
Fig. 2.
The K63 isopeptide Ub linkage-specific mAb, HWA4C4. (A) Purified HWA4C4 (1 μg/mL) was tested by ELISA for reactivity against all 7 branched peptides, U[58–66] peptide, U[68–76] peptide, and Ub protein. (B) The HWA4C4 antibody was immobilized on CM5 biosensor chip and tested by Biacore analysis for binding ability to all 7 branched peptides, U[58–66] peptide, and U[68–76] peptide at the indicated concentrations.
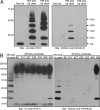
Fig. 3.
HWA4C4 is specific for K63-linked polyUb. (A) Equal amounts (300 ng) of free Ub, K63-linked polyUb, and K48-linked polyUb were separated by SDS/PAGE and immunoblotted with either Ub mAb (P4D1) or K63Ub mAb (HWA4C4). (B) In addition to K63-linked polyUb (300 ng), equal amounts of K6-, K11-, K27-, K29-, K33-, K48-, and K63-linked polyUb-modified substrates (1 μg), ([KX]Ub)_n_-substrate, were separated by SDS/PAGE and immunoblotted with either Ub mAb (P4D1) or K63Ub mAb (HWA4C4).
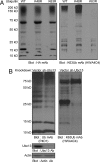
Fig. 4.
HWA4C4 can specifically detect K63-linked polyUb-modified proteins in whole lysates. (A) HEK293T cells were transiently transfected with HA-tagged wild-type (WT), K48R, or K63R Ub. Cells were lysed 48 h after transfection. Cell lysates were gel-separated and immunoblotted with HA and K63Ub antibodies. (B) The lysates of stably transfected A20 cells expressing shRNA targeting Ubc13 (37) or an empty vector control were gel-separated and immunoblotted with Ub, K63Ub, Ubc13, and actin antibodies.
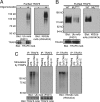
Fig. 5.
Ex vivo analysis of TRAF6 polyubiquitylation. (A) TAP-tagged TRAF6-gyrase B in a stable HEK293T cell line was activated by oligomerization with coumermycin A. TRAF6 was TAP-purified from lysates of cells incubated with or without coumermycin A, gel-separated, and immunoblotted with P4D1 and HWA4C4. (B) WT and RING-mutated (MURING) TAP-tagged TRAF6 constructs were overexpressed in HEK293T cells, TAP-purified, and gel-separated. Blots were probed with P4D1 and HWA4C4. (C) (Upper) RAW264.7 cells were stimulated with LPS (1 μg/mL), CpG DNA (1 μM), and poly(I·C) (10 μg/mL) for 10 min. TRAF6 and TRAF2 were immunoprecipitated from cell lysates, gel-separated, and immunoblotted with P4D1 or HWA4C4. (Lower) Comparable loading was confirmed by reprobing with TRAF6 and TRAF2 antibodies.
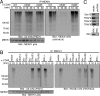
Fig. 6.
Ex vivo analysis of MEKK1 polyubiquitylation. (A) (Upper) Splenic CD43− B cells were stimulated with an agonistic CD40 antibody, and at the indicated times MEKK1 was immunoprecipitated and washed extensively. In vitro ubiquitylation assays were performed on the immunoprecipitated MEKK1 (which is free of TRAF2) with E1 and E2 Ub-conjugating enzymes plus WT Ub or Ub mutants (K63R or K48R). The reaction mixtures were separated by SDS/PAGE, and blots were probed with P4D1 or HWA4C4. (Lower) Comparable loading of MEKK1 was confirmed by reprobing with a MEKK1 Ab. (B) (Upper) Stably transfected A20 cells expressing shRNA targeting Ubc13, TRAF2, TRAF3, TRAF6, or an empty vector control were stimulated with CD40 Ab for the times indicated. The B cell lines 1.3E2 (_Ikk_γ−) and 70Z3 (_Ikk_γ+) were also stimulated with CD40 Ab. MEKK1 was immunoprecipitated, gel-separated, and immunoblotted with P4D1 or HWA4C4. (Lower) Comparable loading was confirmed by reprobing the same blots with MEKK1 antibody. (C) Appropriate knockdowns or absence of IKKγ in the cell lines examined above were confirmed by immunoblotting.
Similar articles
- Using linkage-specific monoclonal antibodies to analyze cellular ubiquitylation.
Newton K, Matsumoto ML, Ferrando RE, Wickliffe KE, Rape M, Kelley RF, Dixit VM. Newton K, et al. Methods Mol Biol. 2012;832:185-96. doi: 10.1007/978-1-61779-474-2_13. Methods Mol Biol. 2012. PMID: 22350886 - Exploring the linkage dependence of polyubiquitin conformations using molecular modeling.
Fushman D, Walker O. Fushman D, et al. J Mol Biol. 2010 Jan 29;395(4):803-14. doi: 10.1016/j.jmb.2009.10.039. Epub 2009 Oct 22. J Mol Biol. 2010. PMID: 19853612 Free PMC article. - Analysis of the biochemical role of Lys-11 in polyubiquitin chain formation using quantitative mass spectrometry.
Jung JW, Bae SJ, Kang GY, Kim KH, Yeo WS, Park SH, Seol JH, Yi EC, Kim KP. Jung JW, et al. Rapid Commun Mass Spectrom. 2013 Jan 30;27(2):339-46. doi: 10.1002/rcm.6447. Rapid Commun Mass Spectrom. 2013. PMID: 23239382 - Atypical Ubiquitination and Parkinson's Disease.
Buneeva O, Medvedev A. Buneeva O, et al. Int J Mol Sci. 2022 Mar 28;23(7):3705. doi: 10.3390/ijms23073705. Int J Mol Sci. 2022. PMID: 35409068 Free PMC article. Review. - [Advances in the application of affinity separation for analyzing protein ubiquitination].
Zhong H, Huang Y, Jin Y, Zhao R. Zhong H, et al. Se Pu. 2021 Jan;39(1):26-33. doi: 10.3724/SP.J.1123.2020.07005. Se Pu. 2021. PMID: 34227356 Free PMC article. Review. Chinese.
Cited by
- Targeting of EGFR by a combination of antibodies mediates unconventional EGFR trafficking and degradation.
Jones S, King PJ, Antonescu CN, Sugiyama MG, Bhamra A, Surinova S, Angelopoulos N, Kragh M, Pedersen MW, Hartley JA, Futter CE, Hochhauser D. Jones S, et al. Sci Rep. 2020 Jan 20;10(1):663. doi: 10.1038/s41598-019-57153-9. Sci Rep. 2020. PMID: 31959764 Free PMC article. - Regulation of immune responses by E3 ubiquitin ligase Cbl-b.
Tang R, Langdon WY, Zhang J. Tang R, et al. Cell Immunol. 2019 Jun;340:103878. doi: 10.1016/j.cellimm.2018.11.002. Epub 2018 Nov 7. Cell Immunol. 2019. PMID: 30442330 Free PMC article. Review. - A putative SUMO interacting motif in the B30.2/SPRY domain of rhesus macaque TRIM5α important for NF-κB/AP-1 signaling and HIV-1 restriction.
Nepveu-Traversy MÉ, Demogines A, Fricke T, Plourde MB, Riopel K, Veillette M, Diaz-Griffero F, Sawyer SL, Berthoux L. Nepveu-Traversy MÉ, et al. Heliyon. 2016 Jan 21;2(1):e00056. doi: 10.1016/j.heliyon.2015.e00056. eCollection 2016 Jan. Heliyon. 2016. PMID: 27441239 Free PMC article. - Generation of a lentiviral vector producer cell clone for human Wiskott-Aldrich syndrome gene therapy.
Wielgosz MM, Kim YS, Carney GG, Zhan J, Reddivari M, Coop T, Heath RJ, Brown SA, Nienhuis AW. Wielgosz MM, et al. Mol Ther Methods Clin Dev. 2015 Jan 21;2:14063. doi: 10.1038/mtm.2014.63. eCollection 2015. Mol Ther Methods Clin Dev. 2015. PMID: 26052531 Free PMC article. - Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis.
Wauer T, Swatek KN, Wagstaff JL, Gladkova C, Pruneda JN, Michel MA, Gersch M, Johnson CM, Freund SM, Komander D. Wauer T, et al. EMBO J. 2015 Feb 3;34(3):307-25. doi: 10.15252/embj.201489847. Epub 2014 Dec 19. EMBO J. 2015. PMID: 25527291 Free PMC article.
References
- Ciechanover A. Intracellular protein degradation: From a vague idea, through the lysosome and the ubiquitin–proteasome system, and onto human diseases and drug targeting (Nobel lecture) Angew Chem Int Ed Engl. 2005;44:5944–5967. - PubMed
- Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. - PubMed
- Glickman MH, Ciechanover A. The ubiquitin–proteasome proteolytic pathway: Destruction for the sake of construction. Physiol Rev. 2002;82:373–428. - PubMed
- Bloom J, et al. Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell. 2003;115:71–82. - PubMed
- Cadwell K, Coscoy L. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science. 2005;309:127–130. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI043477/AI/NIAID NIH HHS/United States
- AI52199/AI/NIAID NIH HHS/United States
- R01 AI052199/AI/NIAID NIH HHS/United States
- R21 AI052199/AI/NIAID NIH HHS/United States
- R56 AI052199/AI/NIAID NIH HHS/United States
- CA21765/CA/NCI NIH HHS/United States
- P30 CA021765/CA/NCI NIH HHS/United States
- R01 AI043477/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous