The DNA-encoded nucleosome organization of a eukaryotic genome - PubMed (original) (raw)
. 2009 Mar 19;458(7236):362-6.
doi: 10.1038/nature07667. Epub 2008 Dec 17.
Affiliations
- PMID: 19092803
- PMCID: PMC2658732
- DOI: 10.1038/nature07667
The DNA-encoded nucleosome organization of a eukaryotic genome
Noam Kaplan et al. Nature. 2009.
Abstract
Nucleosome organization is critical for gene regulation. In living cells this organization is determined by multiple factors, including the action of chromatin remodellers, competition with site-specific DNA-binding proteins, and the DNA sequence preferences of the nucleosomes themselves. However, it has been difficult to estimate the relative importance of each of these mechanisms in vivo, because in vivo nucleosome maps reflect the combined action of all influencing factors. Here we determine the importance of nucleosome DNA sequence preferences experimentally by measuring the genome-wide occupancy of nucleosomes assembled on purified yeast genomic DNA. The resulting map, in which nucleosome occupancy is governed only by the intrinsic sequence preferences of nucleosomes, is similar to in vivo nucleosome maps generated in three different growth conditions. In vitro, nucleosome depletion is evident at many transcription factor binding sites and around gene start and end sites, indicating that nucleosome depletion at these sites in vivo is partly encoded in the genome. We confirm these results with a micrococcal nuclease-independent experiment that measures the relative affinity of nucleosomes for approximately 40,000 double-stranded 150-base-pair oligonucleotides. Using our in vitro data, we devise a computational model of nucleosome sequence preferences that is significantly correlated with in vivo nucleosome occupancy in Caenorhabditis elegans. Our results indicate that the intrinsic DNA sequence preferences of nucleosomes have a central role in determining the organization of nucleosomes in vivo.
Figures
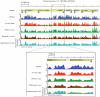
Figure 1. The intrinsic DNA-encoded nucleosome organization at a typical genomic region
Shown are the four different maps of nucleosome occupancy measured in this study for a typical 20,000-bp-long genomic region: the in vitro map, which reflects only the intrinsic nucleosome sequence preferences, and in vivo yeast maps for three different growth conditions (YPD, ethanol and galactose). Each track plots the measured nucleosome occupancy per base pair, computed by summing all of the nucleosome reads obtained in that experiment, and dividing that number by the average number of reads per base pair across the genome. The line of y = 1 thus represents the genome-wide average and is shown as a dashed orange line. The average nucleosome occupancy predictions from our model are shown in blue.
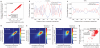
Figure 2. In vitro and in vivo maps are highly similar
a, Shown is a density dot plot comparison of the normalized nucleosome occupancy per base pair in the in vitro (x axis) and in vivo (y axis) maps (see Methods). Values above zero indicate nucleosome enrichment relative to the genome-wide average. The colour of each point represents the number of base pairs that map to that point in the graph. The Pearson correlation between the maps is indicated. b, Nucleosome depletion in vivo relative to in vitro over coding regions increases with the expression level of associated genes. Shown is a dot plot comparison between the expression level of every yeast gene (measured in ref. 26) and the difference between the average normalized nucleosome occupancy of the coding region of that gene in the in vitro map compared with the in vivo map (that is, higher values indicate larger nucleosome depletion in vivo relative to in vitro). The Pearson correlation of the dot plot is indicated.
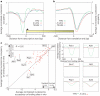
Figure 3. The in vitro sequence preferences of nucleosomes are highly similar to those of nucleosome-bound sequences in vivo and are predictive of nucleosome occupancy in C. elegans
a, Comparison of genome-wide relative nucleosome occupancy of nucleosomes over sequences of length 5. For the in vitro and in vivo maps of nucleosome occupancy, we separately computed the average normalized nucleosome occupancy of each of the 1,024 sequences of length 5, across all of its instances in the genome. Shown is a comparison between the distributions of these 5-base-pair sequences in both maps. Also shown is the Pearson correlation between these distributions. b, Position-dependent sequence preferences of nucleosomes in the in vitro map. We aligned the individual nucleosome reads in the in vitro nucleosome collection. Shown is the fraction (3-bp moving average) of AA/AT/TT/TA and CC/CG/GC/GG dinucleotides at each position of the alignment. c, Same as b, for the in vivo map. d, Shown is a density dot plot comparison between the normalized nucleosome occupancy per base pair in the in vitro map (x axis) and the normalized nucleosome occupancy per base pair predicted by our cross-validated computational model of nucleosome sequence preferences (y axis). Values above zero indicate nucleosome enrichment relative to the genome-wide average. The colour of each point represents the number of base pairs that map to that point in the graph. The Pearson correlation between the maps is indicated. e, Same as d, comparing our model predictions to the in vivo map. f, In vitro nucleosome sequence preferences on yeast genomic DNA are predictive of the in vivo nucleosome organization in C. elegans. Same as d, comparing our model predictions and the in vivo nucleosome occupancy map of C. elegans on chromosome 2 (ref. 19). g, Comparison of yeast nucleosome sequence preferences in vitro and those of C. elegans in vivo. For each of the maps we separately computed the average normalized nucleosome occupancy of every possible sequence of length 5. For C. elegans, we performed these computations on chromosome 2. Shown is a comparison of these 5-base-pair sequence distributions between the yeast in vitro map and the in vivo map of C. elegans, along with the Pearson correlation between these distributions.
Figure 4. The intrinsic nucleosome organization over transcripts and transcription factor binding sites
a, For the in vitro and in vivo nucleosome occupancy maps, and for our model, shown is the normalized nucleosome occupancy per base pair around the transcription start site, averaged across all yeast genes. The long-range ordering of nucleosome occupancy which is present in the in vivo maps but not in the in vitro map may be partly explained by the lower nucleosome concentration in which the in vitro experiment was carried out (see Methods), because higher nucleosome concentration in vivo is predicted to cause long-range ordering of nucleosome arrays. b, Same as a, but around translation end sites of genes (translation end was chosen because transcription end sites are poorly annotated). The depletion around gene ends may be due to the presence of termination signals, which disfavour nucleosome formation in vitro (Supplementary Fig. 8). The fact that these signals tend to occur in a specific orientation with respect to the direction of transcription is consistent with a function in transcript processing, but does not exclude the possibility that one or more of these motifs functions primarily to disfavour nucleosomes. c, Comparison of the nucleosome occupancy over transcription factor binding sites between the in vitro and the YPD in vivo maps. For each transcription factor with at least 50 functional binding sites, we computed, separately for the in vivo and in vitro maps, the average normalized nucleosome occupancy over its binding sites. Shown is a comparison of these nucleosome occupancies per factor, between the in vivo and in vitro maps, along with the Pearson correlation between them. For six factors taken from different regions of the plot, we also show the average normalized nucleosome occupancy around those factors’ binding sites, for both the in vitro and the in vivo maps.
Similar articles
- A genomic code for nucleosome positioning.
Segal E, Fondufe-Mittendorf Y, Chen L, Thåström A, Field Y, Moore IK, Wang JP, Widom J. Segal E, et al. Nature. 2006 Aug 17;442(7104):772-8. doi: 10.1038/nature04979. Epub 2006 Jul 19. Nature. 2006. PMID: 16862119 Free PMC article. - Diversity of eukaryotic DNA replication origins revealed by genome-wide analysis of chromatin structure.
Berbenetz NM, Nislow C, Brown GW. Berbenetz NM, et al. PLoS Genet. 2010 Sep 2;6(9):e1001092. doi: 10.1371/journal.pgen.1001092. PLoS Genet. 2010. PMID: 20824081 Free PMC article. - Prediction of nucleosome positioning in genomes: limits and perspectives of physical and bioinformatic approaches.
De Santis P, Morosetti S, Scipioni A. De Santis P, et al. J Biomol Struct Dyn. 2010 Jun;27(6):747-64. doi: 10.1080/07391102.2010.10508583. J Biomol Struct Dyn. 2010. PMID: 20232931 - Nucleosome positioning in yeasts: methods, maps, and mechanisms.
Lieleg C, Krietenstein N, Walker M, Korber P. Lieleg C, et al. Chromosoma. 2015 Jun;124(2):131-51. doi: 10.1007/s00412-014-0501-x. Epub 2014 Dec 23. Chromosoma. 2015. PMID: 25529773 Review. - Active nucleosome positioning beyond intrinsic biophysics is revealed by in vitro reconstitution.
Korber P. Korber P. Biochem Soc Trans. 2012 Apr;40(2):377-82. doi: 10.1042/BST20110730. Biochem Soc Trans. 2012. PMID: 22435815 Review.
Cited by
- Quality control of transcription start site selection by nonsense-mediated-mRNA decay.
Malabat C, Feuerbach F, Ma L, Saveanu C, Jacquier A. Malabat C, et al. Elife. 2015 Apr 23;4:e06722. doi: 10.7554/eLife.06722. Elife. 2015. PMID: 25905671 Free PMC article. - ENCODE: A Sourcebook of Epigenomes and Chromatin Language.
Yavartanoo M, Choi JK. Yavartanoo M, et al. Genomics Inform. 2013 Mar;11(1):2-6. doi: 10.5808/GI.2013.11.1.2. Epub 2013 Mar 31. Genomics Inform. 2013. PMID: 23613676 Free PMC article. - NucleoFinder: a statistical approach for the detection of nucleosome positions.
Becker J, Yau C, Hancock JM, Holmes CC. Becker J, et al. Bioinformatics. 2013 Mar 15;29(6):711-6. doi: 10.1093/bioinformatics/bts719. Epub 2013 Jan 6. Bioinformatics. 2013. PMID: 23297036 Free PMC article. - Perfect and imperfect nucleosome positioning in yeast.
Cole HA, Nagarajavel V, Clark DJ. Cole HA, et al. Biochim Biophys Acta. 2012 Jul;1819(7):639-43. doi: 10.1016/j.bbagrm.2012.01.008. Epub 2012 Jan 28. Biochim Biophys Acta. 2012. PMID: 22306662 Free PMC article. Review. - Promoter nucleosome organization shapes the evolution of gene expression.
Rosin D, Hornung G, Tirosh I, Gispan A, Barkai N. Rosin D, et al. PLoS Genet. 2012;8(3):e1002579. doi: 10.1371/journal.pgen.1002579. Epub 2012 Mar 15. PLoS Genet. 2012. PMID: 22438828 Free PMC article.
References
- Feng HP, Scherl DS, Widom J. Lifetime of the histone octamer studied by continuous-flow quasielastic light scattering: test of a model for nucleosome transcription. Biochemistry. 1993;32:7824–7831. - PubMed
- Porreca GJ, et al. Multiplex amplification of large sets of human exons. Nature Methods. 2007;4:931–936. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA119176/CA/NCI NIH HHS/United States
- R01 GM072518-02/GM/NIGMS NIH HHS/United States
- R01 GM072518-04/GM/NIGMS NIH HHS/United States
- R01 GM072518-03/GM/NIGMS NIH HHS/United States
- R01 GM058617/GM/NIGMS NIH HHS/United States
- R01 GM058617-11/GM/NIGMS NIH HHS/United States
- R01 GM054692/GM/NIGMS NIH HHS/United States
- R01 GM054692-11/GM/NIGMS NIH HHS/United States
- R01 GM072518/GM/NIGMS NIH HHS/United States
- R01 GM072518-01A1/GM/NIGMS NIH HHS/United States
- R01 CA119176-03/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases