Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex - PubMed (original) (raw)
Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex
Yanli Wang et al. Nature. 2008.
Abstract
Here we report on a 3.0 A crystal structure of a ternary complex of wild-type Thermus thermophilus argonaute bound to a 5'-phosphorylated 21-nucleotide guide DNA and a 20-nucleotide target RNA containing cleavage-preventing mismatches at the 10-11 step. The seed segment (positions 2 to 8) adopts an A-helical-like Watson-Crick paired duplex, with both ends of the guide strand anchored in the complex. An arginine, inserted between guide-strand bases 10 and 11 in the binary complex, locking it in an inactive conformation, is released on ternary complex formation. The nucleic-acid-binding channel between the PAZ- and PIWI-containing lobes of argonaute widens on formation of a more open ternary complex. The relationship of structure to function was established by determining cleavage activity of ternary complexes containing position-dependent base mismatch, bulge and 2'-O-methyl modifications. Consistent with the geometry of the ternary complex, bulges residing in the seed segments of the target, but not the guide strand, were better accommodated and their complexes were catalytically active.
Figures
Figure 1. Crystal structure of T. thermophilus Ago bound to 5′-phosphorylated 21-nucleotide guide DNA and 20-nucleotide target RNA
a, Sequence of the guide DNA-target RNA duplex. The traceable segments of the bases of the guide DNA and target RNA in the structure of the ternary complex are shown in red and blue, respectively. Disordered segments of the bases on both strands that cannot be traced are shown in grey. b, Stereo view of the 3.0 Å crystal structure of the Ago ternary complex. The Ago protein is colour-coded by domains (N in cyan, PAZ in magenta, Mid in orange and PIWI in green) and linkers (L1 and L2 in grey). The bound 21-nucleotide guide DNA is in red and traced for bases of the 1–10 and 19–21 segments, whereas the bound 20-nucleotide target RNA is in blue and traced for bases of the 1′ to 9′ segment. Backbone phosphorus atoms are in yellow. c, An alternate view of the complex.
Figure 2. Comparison of structural details between the binary Ago complex with bound guide DNA and the ternary complex with added target RNA
a, Expanded view of the ternary complex highlighting the guide DNA (1–10)-target RNA (1′–9′) duplex and Mg2+-coordinated catalytic residues (D478, D546 and D660) of the RNase H fold of the PIWI domain. Intermolecular hydrogen bonds between the Ago protein and the DNA guide strand in red are shown by dashed lines. b, Positioning of the sugar-phosphate backbone of the target RNA strand spanning the mismatch-containing 10–11-step relative to the catalytic residues of the PIWI domain. c, Comparison of the trajectory of traceable bound guide DNA in the binary (bases 1–11 and 18–21 in silver) and ternary (bases 1–10 and 19–21 in red) Ago complexes after superposition of their 5′-phosphate-binding pockets. d, Superposition of the guide DNA (red)-target RNA (blue) duplex spanning the 2–8 seed segment on A-form (left panel) and B-form (right panel) helices (silver) after best-fit superposition of the target RNA strand of the ternary Ago complex with one strand of the A/B-form helices. e, Positioning of stacked residues 6–10 of the DNA guide strand relative to R548, with emphasis on intermolecular interactions involving the sugar-phosphate backbone. f, Relative positioning of the 6 to 10/11 segment of the bound guide DNA strand and R548 in the binary (guide strand in silver, protein in cyan) and ternary (guide strand in red, protein in magenta) Ago complexes. The conformational change in the protein on proceeding from binary to ternary Ago complexes is indicated by a red arrow.
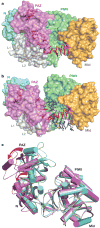
Figure 3. Conformational changes within the bilobal Ago scaffold on proceeding from the binary (guide) complex to the ternary (guide + target) complex
a, b, The nucleic acid binding canyon changes width on proceeding from the binary (a) to the ternary (b) complex. The Ago protein is shown in a space-filling representation with labelled domains and linkers colour-coded as in Fig. 1. The guide DNA (red) and target RNA (blue) are shown in stick representation with backbone phosphorus atoms in yellow. c, View of alignment of binary (cyan) and ternary (magenta) Ago complexes, after superpositioning of their PIWI-containing modules. The red arrows indicate the magnitude of the conformational changes on proceeding from binary to ternary complexes.
Figure 4. Target RNA cleavage activity of T. thermophilus Ago loaded with mismatched, bulge-containing or length-altered guide DNA strands
Besides Ago-mediated target RNA cleavage, extensive chemical hydrolysis is observed over the entire RNA substrate length, except for a 12-nucleotide (nt) region across guide DNA positions 2 to 13, which was protected from hydrolysis by the interaction with Ago and its guide DNA. Mismatches or bulges in the 5′ region reduce protection, and frequently catalysis, because of reduced base-pairing stability and deviation from hydrolysis-protecting helical geometry. The black bar to the left of the images defines the region of the cleavage substrate complementary with the 21-nucleotide (nt) guide DNA (a–d) or the region complementary with the 7-nucleotide guide DNA with the dotted line indicating pairing for 3′-extended guide DNAs (e). Arrows indicate the cleavage site. Del, deletion; H, hydrolysis ladder of substrate RNA; T1, partial RNase T1 digest of substrate RNA. a, Single mismatches were placed in the 21-nucleotide guide, at positions 1 to 19. b, Double, quadruple, sextuple and octuple mismatches were introduced in the DNA guide 3′ region at indicated positions. c, Cleavage assay using DNA guides with a deletion or insertions of bulged nucleotides at indicated positions. d, Insertion of bulges in the target RNA. Insertion of nucleotides upstream of the cleavage site leads to shifts in the cleavage site by the number of inserted nucleotides. e, DNA guide length was increased from 7 to 36 nucleotides. The sequences of DNA guides and RNA targets are listed in Supplementary Table 2. As evident from increased hydrolysis of the target region recognized by the guide, insertion of bulges seems to destabilize guide-target interactions.
Similar articles
- Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes.
Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ. Wang Y, et al. Nature. 2009 Oct 8;461(7265):754-61. doi: 10.1038/nature08434. Nature. 2009. PMID: 19812667 Free PMC article. - Structure of the guide-strand-containing argonaute silencing complex.
Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Wang Y, et al. Nature. 2008 Nov 13;456(7219):209-13. doi: 10.1038/nature07315. Epub 2008 Aug 27. Nature. 2008. PMID: 18754009 Free PMC article. - Structure/cleavage-based insights into helical perturbations at bulge sites within T. thermophilus Argonaute silencing complexes.
Sheng G, Gogakos T, Wang J, Zhao H, Serganov A, Juranek S, Tuschl T, Patel DJ, Wang Y. Sheng G, et al. Nucleic Acids Res. 2017 Sep 6;45(15):9149-9163. doi: 10.1093/nar/gkx547. Nucleic Acids Res. 2017. PMID: 28911094 Free PMC article. - Molecular mechanism of target RNA transcript recognition by Argonaute-guide complexes.
Parker JS, Roe SM, Barford D. Parker JS, et al. Cold Spring Harb Symp Quant Biol. 2006;71:45-50. doi: 10.1101/sqb.2006.71.029. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381279 Review. - Understanding the core of RNA interference: The dynamic aspects of Argonaute-mediated processes.
Zhu L, Jiang H, Sheong FK, Cui X, Wang Y, Gao X, Huang X. Zhu L, et al. Prog Biophys Mol Biol. 2017 Sep;128:39-46. doi: 10.1016/j.pbiomolbio.2016.09.008. Epub 2016 Sep 30. Prog Biophys Mol Biol. 2017. PMID: 27697475 Review.
Cited by
- Hsp90 cochaperones p23 and FKBP4 physically interact with hAgo2 and activate RNA interference-mediated silencing in mammalian cells.
Pare JM, LaPointe P, Hobman TC. Pare JM, et al. Mol Biol Cell. 2013 Aug;24(15):2303-10. doi: 10.1091/mbc.E12-12-0892. Epub 2013 Jun 5. Mol Biol Cell. 2013. PMID: 23741051 Free PMC article. - A regulatory loop between miR-132 and miR-125b involved in gonadotrope cells desensitization to GnRH.
Lannes J, L'hôte D, Fernandez-Vega A, Garrel G, Laverrière JN, Cohen-Tannoudji J, Quérat B. Lannes J, et al. Sci Rep. 2016 Aug 19;6:31563. doi: 10.1038/srep31563. Sci Rep. 2016. PMID: 27539363 Free PMC article. - Prokaryotic Argonaute Proteins as a Tool for Biotechnology.
Kropocheva EV, Lisitskaya LA, Agapov AA, Musabirov AA, Kulbachinskiy AV, Esyunina DM. Kropocheva EV, et al. Mol Biol. 2022;56(6):854-873. doi: 10.1134/S0026893322060103. Epub 2022 Aug 30. Mol Biol. 2022. PMID: 36060308 Free PMC article. - Diversifying microRNA sequence and function.
Ameres SL, Zamore PD. Ameres SL, et al. Nat Rev Mol Cell Biol. 2013 Aug;14(8):475-88. doi: 10.1038/nrm3611. Epub 2013 Jun 26. Nat Rev Mol Cell Biol. 2013. PMID: 23800994 Review. - Allosteric regulation of Argonaute proteins by miRNAs.
Djuranovic S, Zinchenko MK, Hur JK, Nahvi A, Brunelle JL, Rogers EJ, Green R. Djuranovic S, et al. Nat Struct Mol Biol. 2010 Feb;17(2):144-50. doi: 10.1038/nsmb.1736. Epub 2010 Jan 10. Nat Struct Mol Biol. 2010. PMID: 20062058 Free PMC article.
References
- Filipowicz W. The nuts and bolts of the RISC machine. Cell. 2005;122:17–20. - PubMed
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nature Rev Mol Cell Biol. 2008;9:22–32. - PubMed
- Tolia NH, Joshua-Tor L. Slicer and the argonautes. Nature Chem Biol. 2007;3:36–43. - PubMed
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources