Cytoplasmic CUG RNA foci are insufficient to elicit key DM1 features - PubMed (original) (raw)
doi: 10.1371/journal.pone.0003968. Epub 2008 Dec 18.
Cordula M Wolf, Partha Sarkar, Sharan Paul, Andy Chiang, Ian Holt, Glenn E Morris, Dorothy Branco, Megan C Sherwood, Lucio Comai, Charles I Berul, Sita Reddy
Affiliations
- PMID: 19092997
- PMCID: PMC2597774
- DOI: 10.1371/journal.pone.0003968
Cytoplasmic CUG RNA foci are insufficient to elicit key DM1 features
Warunee Dansithong et al. PLoS One. 2008.
Abstract
The genetic basis of myotonic dystrophy type I (DM1) is the expansion of a CTG tract located in the 3' untranslated region of DMPK. Expression of mutant RNAs encoding expanded CUG repeats plays a central role in the development of cardiac disease in DM1. Expanded CUG tracts form both nuclear and cytoplasmic aggregates, yet the relative significance of such aggregates in eliciting DM1 pathology is unclear. To test the pathophysiology of CUG repeat encoding RNAs, we developed and analyzed mice with cardiac-specific expression of a beta-galactosidase cassette in which a (CTG)(400) repeat tract was positioned 3' of the termination codon and 5' of the bovine growth hormone polyadenylation signal. In these animals CUG aggregates form exclusively in the cytoplasm of cardiac cells. A key pathological consequence of expanded CUG repeat RNA expression in DM1 is aberrant RNA splicing. Abnormal splicing results from the functional inactivation of MBNL1, which is hypothesized to occur due to MBNL1 sequestration in CUG foci or from elevated levels of CUG-BP1. We therefore tested the ability of cytoplasmic CUG foci to elicit these changes. Aggregation of CUG RNAs within the cytoplasm results both in Mbnl1 sequestration and in approximately a two fold increase in both nuclear and cytoplasmic Cug-bp1 levels. Significantly, despite these changes RNA splice defects were not observed and functional analysis revealed only subtle cardiac dysfunction, characterized by conduction defects that primarily manifest under anesthesia. Using a human myoblast culture system we show that this transgene, when expressed at similar levels to a second transgene, which encodes expanded CTG tracts and facilitates both nuclear focus formation and aberrant splicing, does not elicit aberrant splicing. Thus the lack of toxicity of cytoplasmic CUG foci does not appear to be a consequence of low expression levels. Our results therefore demonstrate that the cellular location of CUG RNA aggregates is an important variable that influences toxicity and support the hypothesis that small molecules that increase the rate of transport of the mutant DMPK RNA from the nucleus into the cytoplasm may significantly improve DM1 pathology.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
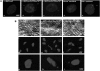
Figure 1. DM1 fibroblasts and myoblasts contain both nuclear and cytoplasmic CUG foci.
Panel A: Nuclear DAPI staining of normal and DM1 fibroblasts and myoblasts, either untreated or immortalized with SV40, are shown in Panels a, d, g, j, m, p, s & v. DMPK transcripts encoding the expanded CUG tracts were detected by hybridization with a (CAG)10-Cy3 probe (red signal; Panels h, k, n, q, t & w). Transcripts containing expanded CUG repeats are not observed in the normal fibroblasts and normal myoblasts (b & e), respectively. Merged images of DAPI and (CAG)10-Cy3 stains show CUG RNA foci as red signals (i, l, o, r, u & x) within the nucleus and in the cytoplasm in DM1 fibroblasts and myoblasts. The percent of DM1 fibroblasts and myoblasts containing both nuclear and cytoplasmic foci are tabulated in Table 1. Images of CUG RNA foci in additional DM1 fibroblasts and myoblasts are shown in supplementary Figure S1 (Panels A–E).
Figure 2. Characterization of α-MHC-LacZ-(CTG)400 mice.
Panel A: The α-MHC-LacZ-(CTG)400 transgene encoding the α-myosin heavy chain promoter (α-MHC) used to drive cardiac specific expression of the β-galactosidase (LacZ) gene followed by a CTG tract of ∼400 repeats and the bovine growth hormone polyA (BGH-PolyA) sequence is shown. Panel B: Restriction digestion of plasmids encoding the α-MHC-LacZ-(CTG)400 sequences with Sfi_I, which allows excision of the CTG repeat tract, demonstrates the instability of the CTG repeats when propagated in E. coli at 37°C. Panel C: Southern blot analysis of mouse tail-clip DNA digested with Pvu_II. The 280 bp probe used for hybridization (Panel A) is shown. The majority of the detected bands contained 350∼380 CTG repeats in α-MHC-LacZ-(CTG)400TG_high (band intensities ∼76%) or 300∼350 CTG repeats in α-MHC-LacZ-(CTG)400TG_low (band intensities ∼80%) in tail clip DNAs. Panel D: Northern blot analysis of RNA derived from α-MHC-LacZ-(CTG)400 and α-MHC-LacZ mouse hearts probed with β-galactosidase and Gapdh sequences is shown.
Figure 3. CUG foci form exclusively in the cytoplasm of α-MHC-LacZ-(CTG)400 cardiomyocytes.
Panel A: Nuclear DAPI staining of cardiomyocytes derived from α-MHC-LacZ, α-MHC-LacZ-(CTG)400 mice and normal human and DM1 myoblasts is shown. Transcripts encoding the expanded CUG tracts were detected by hybridization with a (CAG)10-Cy3 probe (red signal). CUG foci are observed in the cytoplasm in α-MHC-LacZ-(CTG)400 cardiomyocytes (b, c) and in both the cytoplasm and nucleus of DM1 myoblasts (e) (120× magnification). Panel B: Nuclear DAPI staining of heart sections of α-MHC-LacZ, α-MHC-LacZ-(CTG)400TG_high_, and α-MHC-LacZ-(CTG)400TG_low_ are shown. CUG foci are observed in the cytoplasm of both α-MHC-LacZ-(CTG)400TG_high_, and α-MHC-LacZ-(CTG)400TG_low_ heart tissue sections [red signals; e, f (40× magnification) and h, i (120× magnification)]. Transcripts containing expanded repeats are not observed in cardiomyocytes and heart sections of α-MHC-LacZ mice and in normal human myoblasts [Panel A; a, d and Panel B; d, _g_]. >400 cells were examined in both α-MHC-LacZ-(CTG)400 cardiomyocyte preparations and in α-MHC-LacZ-(CTG)400 cardiac sections.
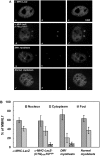
Figure 4. Mbnl1 co-localizes with cytoplasmic CUG-foci in α-MHC-LacZ-(CTG)400 cardiomyocytes.
Panel A: Distribution of endogenous Mbnl1 is visualized as a green signal (a, d, g & j) using anti-MBNL1 (MB1a) monoclonal antibody and secondary antibodies conjugated with FITC. The mutant transcripts encoding the expanded CUG tracts were detected by hybridization with a (CAG)10-Cy3 probe (red signals; e & h). Transcripts containing expanded CUG repeats are not observed in the α-MHC-LacZ cardiomyocytes (b) and normal myoblasts (k). Merged images (f & i), where super-imposition of green and red signals are observed as a yellow signals, show that Mbnl1 co-localizes with the expanded CUG tracts in the α-MHC-LacZ-(CUG)400 cardiomyocytes (f) and in DM1 myoblasts (i). Panel B: Graphical representation of Mbnl1 distribution in each compartment (nucleus, cytoplasm and foci) of α-MHC-LacZ, α-MHC-LacZ-(CUG)400 cardiomyocytes and DM1 and normal myoblasts is shown and the results are tabulated in Table 2. No significant difference is observed in the fraction of Mbnl1 which colocalizes with the foci in α-MHC-LacZ-(CTG)400 cardiomyocytes and in DM1 myoblasts (p = 0.37). The specificity of MBNL1 (MB1a) monoclonal antibody was assessed by immunofluorescence using cardiomyocytes derived from Mbnl1−/− mice (Supplementary Figure S2).
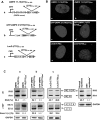
Figure 5. α-MHC-LacZ-(CTG)400 mice show increased steady-state levels of Cug-bp1.
Panels A–B: Protein extracts were prepared from α-MHC-LacZ, α-MHC-LacZ-(CTG)400TG_high_, and α-MHC-LacZ-(CTG)400TG_low_ mouse hearts and 6 or 10 µg of the total proteins from the tissue extracts were resolved on SDS-PAGE followed by Western blot analyses and immunostaining with CUG-BP1 and MBNL1 monoclonal antibodies (mAb), respectively. The blots were re-probed for GAPDH using anti-GAPDH polyclonal antibodies as an internal control. The experiments were carried out in triplicate and mean values of steady-state Cug-bp1 and Mbnl1 levels are shown. Panel C: Cytoplasmic and nuclear proteins extracts (10 µg) from α-MHC-LacZ, α-MHC-LacZ-(CTG)400TG_high_, and α-MHC-LacZ-(CTG)400TG_low_ mouse hearts were resolved on SDS-PAGE followed by Western blot analyses and immunostaining with CUG-BP1 mAb. The blots were re-probed for TATA binding protein (TBP), and for GAPDH, which were used as nuclear and cytoplasmic markers respectively. The experiments were carried out in triplicate and mean values of steady-state Cug-bp1 levels are shown.
Figure 6. Aberrant splicing is not observed in α-MHC-LacZ-(CTG)400 hearts.
Total RNA isolated from adult α-MHC-LacZ-(CTG)400 and adult α-MHC-LacZ hearts and wild-type postnatal day1 and day 2 mouse hearts was subjected to RT-PCR analysis using the Tnnt2, Alp, Zasp and m-Ttn primers as described in Methods. Gapdh RNA was amplified in parallel as an internal control. The experiments were carried out in triplicate and the results are tabulated in Table 3.
Figure 7. Expression of LacZ-(CUG)400 RNAs is insufficient to dysregulate IR and cTNT splicing in human myoblasts.
Panel A: DMPK 11-15(CTG)5 or 300 (a), GFP-DMPK 3′UTR (CTG)5 or 400 (b) and LacZ-(CTG)0 or 400 (c) cassettes under the transcriptional control of the cytomegalovirus (CMV) promoter are shown. Panel B: Nuclear DAPI staining of human normal myoblasts expressing DMPK11-15(CTG)5 (a), DMPK 11-15(CTG)300 (b), GFP-DMPK 3′UTR(CTG)5 (c), GFP-DMPK 3′UTR(CTG)400 (d), LacZ-(CTG)0 (e), LacZ-(CTG)400 (f) cassettes are shown. The mutant transcripts encoding the expanded CUG tracts were detected by hybridization with a (CAG)10-Cy3 probe. CUG RNA foci are observed primarily within the nucleus in normal myoblasts expressing DMPK11-15(CTG)300 (red signal; b) and GFP-DMPK 3′UTR (CTG)400 (red signal; d). CUG RNA foci are observed in the cytoplasm (red signal; f) in normal myoblasts expressing the LacZ-(CTG)400 cassette. Normal myoblasts expressing DMPK11-15(CTG)5 (a), GFP-DMPK 3′UTR(CTG)5 (c), and LacZ-(CTG)0 (e) constructs did not show RNA foci. Panel C: IR and cTNT RNA splicing in myoblasts expressing the indicated cassettes are shown. Synthesized cDNAs (150 ng) were subjected to RT-PCR analysis using the IR and cTNT primers described in Methods. GAPDH RNA was amplified in parallel as an internal control. The experiments were carried out in triplicate. Representative panels are shown in Panel C and the results are tabulated in Table 4.
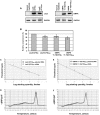
Figure 8. Quantitation of the steady-state levels of LacZ-(CUG)400 and DMPK 11-15(CUG)300 RNAs.
Panels A–B: RT-PCR analyses of the steady-state expression levels of LacZ-(CTG)0, LacZ-(CTG)400, and DMPK11-15(CTG)5, DMPK 11-15(CTG)300 cassettes are shown. Synthesized cDNA (100 ng) from normal myoblsts expressing LacZ-(CTG)0 or 400 and DMPK 11-15(CTG)5 or 300 were subjected to RT-PCR analysis. GAPDH RNA was amplified in parallel as an internal control. The experiments were carried out in triplicate and the results are tabulated in Table 5. Relative steady-state expression levels of the LacZ-(CTG)400 and DMPK 11-15(CTG)300 cassettes were not significantly different (p = 0.479). Panels C–F: Real-time PCR analysis of serial dilutions of plasmid DNAs encoding LacZ-(CTG)400 and DMPK 11-15(CTG)300 sequences and of LacZ-(CTG)400 and DMPK 11-15(CTG)300 cDNAs is shown. PCR reactions were carried using 10−2 to 10−6 fmoles of plasmid DNAs encoding LacZ-(CTG)400 or DMPK 11-15(CTG)300 sequences. To quantitate the expression levels of expanded CUG repeat encoding transcripts, cDNAs (5 ng) from human myoblasts expressing LacZ-(CTG)400 and DMPK 11-15(CTG)300 were subjected to Real-time PCR analysis in parallel. LacZ-(CTG)400 and DMPK 11-15(CTG)300 cDNAs are present at approximately similar levels (Panels C, E & Table 6). Melting curves of LacZ-(CTG)400 or DMPK 11-15(CTG)300 PCR reactions are shown (Panels D & F). GAPDH was used as an internal control for RNA quality and the reverse transcriptase reaction (Ct values for GAPDH in LacZ-(CTG)400 and DMPK 11-15(CTG)300 samples was 19.8 in each case).
Figure 9. Aberrant mitochondrial cristae are observed in α-MHC-LacZ-(CTG)400 hearts.
Electron micrographs of adult heart sections of α-MHC-LacZ (A) and α-MHC-LacZ-(CTG)400TG_high_ (B) mice are shown. Mitochondrial cristae were disarrayed in a subset of sections viewed.
Similar articles
- Elevation of RNA-binding protein CUGBP1 is an early event in an inducible heart-specific mouse model of myotonic dystrophy.
Wang GS, Kearney DL, De Biasi M, Taffet G, Cooper TA. Wang GS, et al. J Clin Invest. 2007 Oct;117(10):2802-11. doi: 10.1172/JCI32308. J Clin Invest. 2007. PMID: 17823658 Free PMC article. - DDX6 regulates sequestered nuclear CUG-expanded DMPK-mRNA in dystrophia myotonica type 1.
Pettersson OJ, Aagaard L, Andrejeva D, Thomsen R, Jensen TG, Damgaard CK. Pettersson OJ, et al. Nucleic Acids Res. 2014 Jun;42(11):7186-200. doi: 10.1093/nar/gku352. Epub 2014 May 3. Nucleic Acids Res. 2014. PMID: 24792155 Free PMC article. - Colocalization of muscleblind with RNA foci is separable from mis-regulation of alternative splicing in myotonic dystrophy.
Ho TH, Savkur RS, Poulos MG, Mancini MA, Swanson MS, Cooper TA. Ho TH, et al. J Cell Sci. 2005 Jul 1;118(Pt 13):2923-33. doi: 10.1242/jcs.02404. Epub 2005 Jun 16. J Cell Sci. 2005. PMID: 15961406 - Gain of RNA function in pathological cases: Focus on myotonic dystrophy.
Klein AF, Gasnier E, Furling D. Klein AF, et al. Biochimie. 2011 Nov;93(11):2006-12. doi: 10.1016/j.biochi.2011.06.028. Epub 2011 Jul 13. Biochimie. 2011. PMID: 21763392 Review. - Molecular mechanisms in DM1 - a focus on foci.
Pettersson OJ, Aagaard L, Jensen TG, Damgaard CK. Pettersson OJ, et al. Nucleic Acids Res. 2015 Feb 27;43(4):2433-41. doi: 10.1093/nar/gkv029. Epub 2015 Jan 20. Nucleic Acids Res. 2015. PMID: 25605794 Free PMC article. Review.
Cited by
- Insights into the mechanism of cell death induced by saporin delivered into cancer cells by an antibody fusion protein targeting the transferrin receptor 1.
Daniels-Wells TR, Helguera G, Rodríguez JA, Leoh LS, Erb MA, Diamante G, Casero D, Pellegrini M, Martínez-Maza O, Penichet ML. Daniels-Wells TR, et al. Toxicol In Vitro. 2013 Feb;27(1):220-31. doi: 10.1016/j.tiv.2012.10.006. Epub 2012 Oct 17. Toxicol In Vitro. 2013. PMID: 23085102 Free PMC article. - Three-dimensional imaging in myotonic dystrophy type 1: Linking molecular alterations with disease phenotype.
Ballester-Lopez A, Núñez-Manchón J, Koehorst E, Linares-Pardo I, Almendrote M, Lucente G, Guanyabens N, Lopez-Osias M, Suárez-Mesa A, Hanick SA, Chojnacki J, Lucia A, Pintos-Morell G, Coll-Cantí J, Martínez-Piñeiro A, Ramos-Fransi A, Nogales-Gadea G. Ballester-Lopez A, et al. Neurol Genet. 2020 Jul 21;6(4):e484. doi: 10.1212/NXG.0000000000000484. eCollection 2020 Aug. Neurol Genet. 2020. PMID: 32802949 Free PMC article. - Repeat-associated non-AUG translation induces cytoplasmic aggregation of CAG repeat-containing RNAs.
Das MR, Chang Y, Anderson R, Saunders RA, Zhang N, Tomberlin CP, Vale RD, Jain A. Das MR, et al. Proc Natl Acad Sci U S A. 2023 Jan 17;120(3):e2215071120. doi: 10.1073/pnas.2215071120. Epub 2023 Jan 9. Proc Natl Acad Sci U S A. 2023. PMID: 36623192 Free PMC article. - Expanded CUG repeats Dysregulate RNA splicing by altering the stoichiometry of the muscleblind 1 complex.
Paul S, Dansithong W, Jog SP, Holt I, Mittal S, Brook JD, Morris GE, Comai L, Reddy S. Paul S, et al. J Biol Chem. 2011 Nov 4;286(44):38427-38438. doi: 10.1074/jbc.M111.255224. Epub 2011 Sep 7. J Biol Chem. 2011. PMID: 21900255 Free PMC article. - RNA imaging in living cells - methods and applications.
Urbanek MO, Galka-Marciniak P, Olejniczak M, Krzyzosiak WJ. Urbanek MO, et al. RNA Biol. 2014;11(8):1083-95. doi: 10.4161/rna.35506. RNA Biol. 2014. PMID: 25483044 Free PMC article. Review.
References
- Harper PS. Myotonic Dystrophy. Third edition, Philadelphia: WB Saunders; 2001.
- Fragola PV, Luzi M, Calo L, Antonini G, Borzi M, et al. Cardiac involvement in myotonic dystrophy. Am J Cardiol. 1994;74:1070–1072. - PubMed
- Hawley RJ, Milner MR, Gottdiener JS, Cohen A. Myotonic heart disease: A clinical follow-up. Neurol. 1991;41:259–262. - PubMed
- Melacini P, Villanova C, Menegazzo E, Novelli G, Danieli G, et al. Correlation between cardiac involvement and CTG trinucleotide repeat length in myotonic dystrophy. J Am Coll Cardiol. 1995;25:239–245. - PubMed
- Tokgozoglu LS, Ashizawa T, Pacifico A, Armstrong RM, Epstein HF, et al. Cardiac involvement in a large kindred with myotonic dystrophy. JAMA. 1995;274:813–819. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS050861/NS/NINDS NIH HHS/United States
- R01 NS060839/NS/NINDS NIH HHS/United States
- 5R01 NS050861/NS/NINDS NIH HHS/United States
- 1R01 NS060839/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources