Allele-specific silencing of mutant Huntington's disease gene - PubMed (original) (raw)
Allele-specific silencing of mutant Huntington's disease gene
Yu Zhang et al. J Neurochem. 2009 Jan.
Abstract
Huntington's disease (HD) is an autosomal-dominant neurodegenerative disorder caused by a poly-glutamine expansion in huntingtin, the protein encoded by the HD gene. PolyQ-expanded huntingtin is toxic to neurons, especially the medium spiny neurons of the striatum. At the same time, wild-type huntingtin has important - indeed essential - protective functions. Any effective molecular therapy must preserve the expression of wild-type huntingtin, while silencing the mutant allele. We hypothesized that an appropriate siRNA molecule would display the requisite specificity and efficacy. As RNA interference is incapable of distinguishing among alleles with varying numbers of CAG (glutamine) codons, another strategy is needed. We used HD fibroblasts in which the pathogenic mutation is linked to a polymorphic site: the Delta2642 deletion of one of four tandem GAG triplets. We silenced expression of the harmful Delta2642-marked polyQ-expanded huntingtin without compromising synthesis of its wild-type counterpart. Following this success in HD fibroblasts, we obtained similar results with neuroblastoma cells expressing both wild-type and mutant HD genes. As opposed to the effect of depleting wild-type huntingtin, specifically silencing the mutant species actually lowered caspase-3 activation and protected HD cells under stress conditions. These findings have therapeutic implications not only for HD, but also for other autosomal dominant diseases. This approach has great promise: it may lead to personalized genetic therapy, a holy grail in contemporary medicine.
Figures
Figure 1
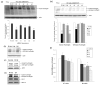
(a) Identification of the Δ2642 triplet deletion in one of four HD fibroblast cells. RT-PCR was conducted using primers spanning the Δ2642 polymorphic locus for three HD fibroblast lines. The reaction products were denatured and resolved in a 6% denaturing TBE-Urea polyacrylamide gel. Lane1; HeLa cells as control; Lane 2: HD fibroblast-GM00305; Lane 3: HD fibroblast-GM06274; Lane 4: HD fibroblast-GM09197. A1 denotes the presence of codon 2642 (112-bp product); A2 denotes the absence of codon 2642 (109-bp product). (b) Sequences of four siRNAs designed to span the polymorphic locus (wild-type or Δ2642).
Figure 2
Allele-specific silencing of endogenous mutant huntingtin. (a) A western blot using 1C2 (MAB1574, Chemicon), an antibody that specifically recognizes mutant huntingtin protein, demonstrates the reduction of mutant huntingtin by siRNAs 2 and 4. (Lysates were prepared 72 hours after transfection. 10μg/lane protein was resolved in 4–20% SDS-PAGE. Note that the MW of huntingtin is approximately 350kDa). A siRNA sequence targeting both the mutant and the wild-type allele (I-htt) was used as a positive control; its reverse sequence (RI-htt) was the negative control. Densitometric analyses of blots from five independent experiments demonstrate that transfection with siRNA of sequence 4 (s4) reduced the level of mutant huntingtin by 43%. The lower part of the membrane was probed with an anti-actin antibody to demonstrate equal loading. Data are presented as the mean ± SEM. Statistical comparisons were made by the Student’s test. HeLa cell lysate was used as a negative control, as these cells only express wild-type huntingtin. * P< 0.05, ** P< 0.01, and *** P< 0.005 as compared to cells transfected with RI-htt. (b) Western blotting using the MAB2166 antibody demonstrates both the reduction of mutant huntingtin and the preservation of wild-type protein in lysates of cells treated with test siRNA s4. An immunoblot of cells treated with I-htt serves as a positive control; one with RI-htt as a negative control. (c) An immunoblot probed with MAB2166 antibody demonstrates that RI-htt (50 nM) doesn’t change levels of either wild-type or mutant huntingtin protein in HD fibroblasts. Control is HD cell transfected without any siRNA. (d) Immunoblots probed with the MAB2166 antibody, and with anti-Bcl-2 and anti-HSP-70 antibodies demonstrated that these two proteins are not affected by siRNA of sequence 4 while there is a significant and specific reduction of mutant huntingtin. (e) A western blot of HD fibroblast lysates was probed with MAB2166 (Chemicon) to determine the effects of the four synthetic siRNAs. HeLa cell lysate was used as a control for wild-type huntingtin. Actin was demonstrated that samples were loading equally. Densitometry was performed on gels from six independent experiments, and signals from huntingtin protein were normalized relative to those from actin. Values for huntingtin in cells transfected with test siRNAs were normalized against ones from cells treated with RI-htt. The data is plotted as the mean ± SEM. * P< 0.05, ** P< 0.01, and *** P< 0.005 as compared to control cells transfected with siRNA of the RI-htt sequence. (f) Real-time qRT-PCR analysis of the endogenous wild-type HD mRNA (wt-allele) and mutant HD mRNA (mu-allele) after transfection with each of the four test siRNAs. HD fibroblasts were transfected with 50 nM of each siRNA for 72 hours and total RNA was harvested for subsequent qRT-PCR analysis. Measurements of the abundance of HD mRNA were normalized relative to signals from GAPDH mRNA in the same sample. Data are reported relative to samples treated with the control sequence. Following transfection with the four test siRNAs, we never observed a significant change in the level of wild-type HD mRNA. By contrast, siRNAs with sequence 2, 3, and 4 all reduced the level of mutant HD mRNA to an appreciable degree, and sequence 4 effected a remarkable 51% drop in the concentration of that message. Data is reported as the average of four independent experiments ± SEM, * P< 0.05, * *P< 0.01, *** P< 0.001 as compared to mutant HD mRNA level after transfection with the control sequence. n = 4.
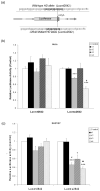
Figure 3
Further confirmation of the specificity of siRNA-mediated silencing of the mutant HD allele using a luciferase reporter assay. (a) Sequences of the inserts present in the luciferase reporter constructs. The first is a 50 bp fragment from the “wild-type” HD allele having codon 2642 and thus four tandem GAG triplets (Lucwt2642); the second is a 47 bp fragment from the “mutant” HD DNA lacking codon 2642 and thus having three GAGs (Lucmu2642). Like the immunoblotting techniques described above, the luciferase-based system was used to measure gene silencing by siRNAs. (b) The Lucwt2642 or Lucmu2642 DNA construct was transfected into HeLa cells together with 20 nM siRNA. Luciferase assays were performed 40 hours after transfection. The signal from luciferase fused to the control sequence in the Luc-wt2642 construct was not diminished by any of the four test siRNAs. By contrast, trasfection with siRNA of sequence 2 or 4 decreased the signal from cells with the Lucmu2642 plasmid. This change in light emission was statistically significant (*** P<0.001, _n_= 6). Only sequence 4 siRNA showed allele-specific inhibition of luciferase expression, i.e., a statistically significant difference in the signals from the Lucwt2642 or Lucmu2642 systems. (# indicates this comparison. P<0.05 as compared to luciferase activity in the Lucwt2642 system transfected with the same siRNAs). Data from six distinct experiments are shown with bars indicating standard errors (± SEM). (c) SH-SY5Y neuroblastoma cells containing Lucwt2642 or Lucmu2642 were transfected with 10 nM each of the four test siRNAs. Forty hours after transfection, luciferase activity from the Lucwt2642 system was not diminished by any of the siRNAs. By contrast, when cells with the Lucmu2642 construct were transfected with s2, s3 or s4 siRNA, they showed significantly less luciferase activity than did the negative control (RT-htt). (*P<0.05, *** P<0.001, _n_= 3). Sequence 4 siRNA exhibited the greatest degree of selectivity in its inhibition of light emission from the Lucmu2642 system relative to that from Lucwt2642 (# P<0.05, # # P<0.01 as compared to luciferase activity from cells with Lucwt2642 that had been transfected with same siRNA sequence). The results from three independent experiments are plotted with bars indicating standard error mean (± SEM).
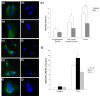
Figure 4
Allele-specific silencing of endogenous mutant huntingtin by siRNA reduced the frequency of HD fibroblasts with abnormal nuclear morphology. It also reduced the extent to which treatment with H2O2 induced caspase-3 activation. (a–j) Control human fibroblasts and HD fibroblasts were stained with Hoechst 33342 dye to reveal nuclear morphology. In contrast to the regular nuclear shape in control cells (a–d), HD cells frequently exhibited nuclear atypia (e–f) and fragmentation (g–j). Sequence 4 siRNA reduced the fraction of HD cells with abnormally shaped and fragmented nuclei from 24 ± 2% to 14 ± 1% (k). The percentage of positive cells was obtained 6–7 different viewings of 200 cells. Cells were counted from a total of seven slides from four independent experiments, *p<0.05, **p< 0.01 as compared to RI-htt transfected controls. (Counting was performed by an investigator blinded to the treatment conditions). (l) Caspase-3-like activity was measured in lysates of H2O2–treated or naïve cells by the hydrolysis of DEVD-AFC. Before exposure to H2O2 (or mock treatment), cells were transfected with siRNA molecules RI-htt, I-htt, or s4. Even when not exposed to 100 μM H2O2, caspase-3 activity increased following transfection with I-htt. (No such effect was observed following transfection with s4 siRNA.) Fifty-six hours after transfection with RI-htt, I-htt or s4, 100 μM of H2O2 was added for an additional 16 hours. In all cases, caspase-3 activity increased. The greatest increase was observed among HD fibroblasts transfected with I-htt; the least with cell treated with s4. * P< 0.05 as compared to cells transfected with RI-htt. _n_=6.
Similar articles
- Increased Steady-State Mutant Huntingtin mRNA in Huntington's Disease Brain.
Liu W, Chaurette J, Pfister EL, Kennington LA, Chase KO, Bullock J, Vonsattel JP, Faull RL, Macdonald D, DiFiglia M, Zamore PD, Aronin N. Liu W, et al. J Huntingtons Dis. 2013;2(4):491-500. doi: 10.3233/JHD-130079. J Huntingtons Dis. 2013. PMID: 25062733 - Inhibition of mutant huntingtin expression by RNA duplex targeting expanded CAG repeats.
Fiszer A, Mykowska A, Krzyzosiak WJ. Fiszer A, et al. Nucleic Acids Res. 2011 Jul;39(13):5578-85. doi: 10.1093/nar/gkr156. Epub 2011 Mar 22. Nucleic Acids Res. 2011. PMID: 21427085 Free PMC article. - Wild type Huntingtin reduces the cellular toxicity of mutant Huntingtin in mammalian cell models of Huntington's disease.
Ho LW, Brown R, Maxwell M, Wyttenbach A, Rubinsztein DC. Ho LW, et al. J Med Genet. 2001 Jul;38(7):450-2. doi: 10.1136/jmg.38.7.450. J Med Genet. 2001. PMID: 11432963 Free PMC article. - Using non-coding small RNAs to develop therapies for Huntington's disease.
Zhang Y, Friedlander RM. Zhang Y, et al. Gene Ther. 2011 Dec;18(12):1139-49. doi: 10.1038/gt.2011.170. Gene Ther. 2011. PMID: 22158031 Review. - The selective vulnerability of nerve cells in Huntington's disease.
Sieradzan KA, Mann DM. Sieradzan KA, et al. Neuropathol Appl Neurobiol. 2001 Feb;27(1):1-21. doi: 10.1046/j.0305-1846.2001.00299.x. Neuropathol Appl Neurobiol. 2001. PMID: 11298997 Review.
Cited by
- A Comprehensive Haplotype-Targeting Strategy for Allele-Specific HTT Suppression in Huntington Disease.
Kay C, Collins JA, Caron NS, Agostinho LA, Findlay-Black H, Casal L, Sumathipala D, Dissanayake VHW, Cornejo-Olivas M, Baine F, Krause A, Greenberg JL, Paiva CLA, Squitieri F, Hayden MR. Kay C, et al. Am J Hum Genet. 2019 Dec 5;105(6):1112-1125. doi: 10.1016/j.ajhg.2019.10.011. Epub 2019 Nov 7. Am J Hum Genet. 2019. PMID: 31708117 Free PMC article. - Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs.
Hu J, Matsui M, Gagnon KT, Schwartz JC, Gabillet S, Arar K, Wu J, Bezprozvanny I, Corey DR. Hu J, et al. Nat Biotechnol. 2009 May;27(5):478-84. doi: 10.1038/nbt.1539. Epub 2009 May 3. Nat Biotechnol. 2009. PMID: 19412185 Free PMC article. - Nucleic Acid-Based Therapy Approaches for Huntington's Disease.
Vagner T, Young D, Mouravlev A. Vagner T, et al. Neurol Res Int. 2012;2012:358370. doi: 10.1155/2012/358370. Epub 2012 Jan 11. Neurol Res Int. 2012. PMID: 22288011 Free PMC article. - CAG expansion in the Huntington disease gene is associated with a specific and targetable predisposing haplogroup.
Warby SC, Montpetit A, Hayden AR, Carroll JB, Butland SL, Visscher H, Collins JA, Semaka A, Hudson TJ, Hayden MR. Warby SC, et al. Am J Hum Genet. 2009 Mar;84(3):351-66. doi: 10.1016/j.ajhg.2009.02.003. Epub 2009 Feb 26. Am J Hum Genet. 2009. PMID: 19249009 Free PMC article. - Influence of mismatched and bulged nucleotides on SNP-preferential RNase H cleavage of RNA-antisense gapmer heteroduplexes.
Magner D, Biala E, Lisowiec-Wachnicka J, Kierzek R. Magner D, et al. Sci Rep. 2017 Oct 2;7(1):12532. doi: 10.1038/s41598-017-12844-z. Sci Rep. 2017. PMID: 28970564 Free PMC article.
References
- Abdelgany A, Wood M, Beeson D. Allele-specific silencing of a pathogenic mutant acetylcholine receptor subunit by RNA interference. Hum Mol Genet. 2003;12:2637–2644. - PubMed
- Almqvist E, Spence N, Nichol K, Andrew SE, Vesa J, Peltonen L, Anvret M, Goto J, Kanazawa I, Goldberg YP, et al. Ancestral differences in the distribution of the delta 2642 glutamic acid polymorphism is associated with varying CAG repeat lengths on normal chromosomes: insights into the genetic evolution of Huntington disease. Hum Mol Genet. 1995;4:207–214. - PubMed
- Ambrose CM, Duyao MP, Barnes G, Bates GP, Lin CS, Srinidhi J, Baxendale S, Hummerich H, Lehrach H, Altherr M, et al. Structure and expression of the Huntington’s disease gene: evidence against simple inactivation due to an expanded CAG repeat. Somat Cell Mol Genet. 1994;20:27–38. - PubMed
- Cattaneo E, Zuccato C, Tartari M. Normal huntingtin function: an alternative approach to Huntington’s disease. Nat Rev Neurosci. 2005;6:919–930. - PubMed
- Difiglia M, Sena-Esteves M, Chase K, Sapp E, Pfister E, Sass M, Yoder J, Reeves P, Pandey RK, Rajeev KG, Manoharan M, Sah DW, Zamore PD, Aronin N. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc Natl Acad Sci U S A. 2007;104:17204–17209. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS051756-05/NS/NINDS NIH HHS/United States
- R01 NS051756/NS/NINDS NIH HHS/United States
- R01 NS039324-09/NS/NINDS NIH HHS/United States
- R01 NS039324/NS/NINDS NIH HHS/United States
- R01 NS039324-08/NS/NINDS NIH HHS/United States
- R01 NS051756-06/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials