Identification of gene 3' ends by automated EST cluster analysis - PubMed (original) (raw)
Identification of gene 3' ends by automated EST cluster analysis
Enrique M Muro et al. Proc Natl Acad Sci U S A. 2008.
Abstract
The properties and biology of mRNA transcripts can be affected profoundly by the choice of alternative polyadenylation sites, making definition of the 3' ends of transcripts essential for understanding their regulation. Here we show that 22-52% of sequences in commonly used human and murine "full-length" transcript databases may not currently end at bona fide polyadenylation sites. To identify probable transcript termini over the entire murine and human genomes, we analyzed the EST databases for positional clustering of EST ends. The analysis yielded 58,282 murine- and 86,410 human-candidate polyadenylation sites, of which 75% mapped to 23,091 known murine transcripts and 22,891 known human transcripts. The murine dataset correctly predicted 97% of the 3' ends in a manually curated and experimentally supported benchmark transcript set. Of currently known genes, 15% had no associated prediction and 25% had only a single predicted termination site. The remaining genes had an average of 3-4 alternative polyadenylation sites predicted for each murine or human transcript, respectively. The results are made available in the form of tables and an interactive web site that can be mined for rapid assessment of the validity of 3' ends in existing collections, enumeration of potential alternative 3' polyadenylation sites of known transcripts, direct retrieval of terminal sequences for design of probes, and detection of polyadenylation sites not currently mapped to known genes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
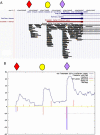
Fig. 1.
EST evidence for alternative 3′ ends for murine Pde7a transcripts. (A) The diagram, obtained from the UCSC Genome Browser (Mouse mm8, February 2006 Assembly) (22), illustrates a region of mouse chromosome 3 spanning 8 kb, including the 3′UTR end of the Pde7a gene, transcribed from right to left. The ends of gene transcript predictions from RefSeq and Ensembl are represented as blue and brown bars, respectively. Black boxes represent the matches to mouse ESTs and mRNAs. Accumulations of EST ends at 3 particular positions are visible (indicated by a red diamond, a yellow oval, and a violet diamond). The position indicated by the red diamond suggests the existence of an alternative termination of the Pde7a gene considered neither by RefSeq nor by Ensembl. (B) Interpretation of EST and PAS information around the predicted 3′UTR of murine Pde7a. The curve, in blue, indicates the number of EST matches at each position. Many of these ESTs end abruptly at the left side of the principal peak, whereas the right side of the peak has a softer slope, which indicates that the ESTs derive from transcripts running from right to left, in agreement with the known direction of transcription of the Pde7a gene. The vertical red lines are maxima of the convolution of the EST-match histogram (see Materials and Methods), which indicate potential terminations. The red lines below the baseline represent potential terminations in the sense of the Pde7a transcription. Further evidence using PAS and clusters of EST ends is then used to confirm transcript ends. The 2 violet vertical bars (under both diamonds) represent clusters of EST ends located near rough ends and composed of at least 2 ESTs ending in the same position with a valid local polyadenylation signal. As explained in A, the rightmost end (violet diamond) is from the Ensembl collection but is found in the RefSeq database; the central end (yellow oval) is represented in Ensembl; and the leftmost (red diamond) is not represented in those collections. Of note, the end marked with the yellow oval had many EST ends (see A). However there was no corresponding PAS, and a tract of 16 consecutive A's coincided with the peak of EST ends. This end, reported by Ensembl, appears to reflect internal transcript priming during cDNA generation rather than a site of transcript termination.
Similar articles
- Discovery of novel human transcript variants by analysis of intronic single-block EST with polyadenylation site.
Wang P, Yu P, Gao P, Shi T, Ma D. Wang P, et al. BMC Genomics. 2009 Nov 12;10:518. doi: 10.1186/1471-2164-10-518. BMC Genomics. 2009. PMID: 19906316 Free PMC article. - Sequence determinants in human polyadenylation site selection.
Legendre M, Gautheret D. Legendre M, et al. BMC Genomics. 2003 Feb 25;4(1):7. doi: 10.1186/1471-2164-4-7. Epub 2003 Feb 25. BMC Genomics. 2003. PMID: 12600277 Free PMC article. - Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data.
Beaudoing E, Gautheret D. Beaudoing E, et al. Genome Res. 2001 Sep;11(9):1520-6. doi: 10.1101/gr.190501. Genome Res. 2001. PMID: 11544195 Free PMC article. - Identification of mRNA polyadenylation sites in genomes using cDNA sequences, expressed sequence tags, and Trace.
Lee JY, Park JY, Tian B. Lee JY, et al. Methods Mol Biol. 2008;419:23-37. doi: 10.1007/978-1-59745-033-1_2. Methods Mol Biol. 2008. PMID: 18369973 - Genome-wide identification and predictive modeling of polyadenylation sites in eukaryotes.
Ji G, Guan J, Zeng Y, Li QQ, Wu X. Ji G, et al. Brief Bioinform. 2015 Mar;16(2):304-13. doi: 10.1093/bib/bbu011. Epub 2014 Apr 1. Brief Bioinform. 2015. PMID: 24695098 Review.
Cited by
- Dual suppression of hemangiogenesis and lymphangiogenesis by splice-shifting morpholinos targeting vascular endothelial growth factor receptor 2 (KDR).
Uehara H, Cho Y, Simonis J, Cahoon J, Archer B, Luo L, Das SK, Singh N, Ambati J, Ambati BK. Uehara H, et al. FASEB J. 2013 Jan;27(1):76-85. doi: 10.1096/fj.12-213835. Epub 2012 Sep 20. FASEB J. 2013. PMID: 22997228 Free PMC article. - All's well that ends well: alternative polyadenylation and its implications for stem cell biology.
Mueller AA, Cheung TH, Rando TA. Mueller AA, et al. Curr Opin Cell Biol. 2013 Apr;25(2):222-32. doi: 10.1016/j.ceb.2012.12.008. Epub 2013 Jan 25. Curr Opin Cell Biol. 2013. PMID: 23357469 Free PMC article. Review. - Construction of mate pair full-length cDNAs libraries and characterization of transcriptional start sites and termination sites.
Matsumoto K, Suzuki A, Wakaguri H, Sugano S, Suzuki Y. Matsumoto K, et al. Nucleic Acids Res. 2014;42(16):e125. doi: 10.1093/nar/gku600. Epub 2014 Jul 17. Nucleic Acids Res. 2014. PMID: 25034687 Free PMC article. - GATExplorer: genomic and transcriptomic explorer; mapping expression probes to gene loci, transcripts, exons and ncRNAs.
Risueño A, Fontanillo C, Dinger ME, De Las Rivas J. Risueño A, et al. BMC Bioinformatics. 2010 Apr 29;11:221. doi: 10.1186/1471-2105-11-221. BMC Bioinformatics. 2010. PMID: 20429936 Free PMC article. - POLYAR, a new computer program for prediction of poly(A) sites in human sequences.
Akhtar MN, Bukhari SA, Fazal Z, Qamar R, Shahmuradov IA. Akhtar MN, et al. BMC Genomics. 2010 Nov 19;11:646. doi: 10.1186/1471-2164-11-646. BMC Genomics. 2010. PMID: 21092114 Free PMC article.
References
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith M, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials