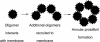Annular protofibrils are a structurally and functionally distinct type of amyloid oligomer - PubMed (original) (raw)
Annular protofibrils are a structurally and functionally distinct type of amyloid oligomer
Rakez Kayed et al. J Biol Chem. 2009.
Abstract
Amyloid oligomers are believed to play causal roles in several types of amyloid-related neurodegenerative diseases. Several different types of amyloid oligomers have been reported that differ in morphology, size, or toxicity, raising the question of the pathological significance and structural relationships between different amyloid oligomers. Annular protofibrils (APFs) have been described in oligomer preparations of many different amyloidogenic proteins and peptides as ring-shaped or pore-like structures. They are interesting because their pore-like morphology is consistent with numerous reports of membrane-permeabilizing activity of amyloid oligomers. Here we report the preparation of relatively homogeneous preparations of APFs and an antiserum selective for APFs (alphaAPF) compared with prefibrillar oligomers (PFOs) and fibrils. PFOs appear to be precursors for APF formation, which form in high yield after exposure to a hydrophobic-hydrophilic interface. Surprisingly, preformed APFs do not permeabilize lipid bilayers, unlike the precursor PFOs. APFs display a conformation-dependent, generic epitope that is distinct from that of PFOs and amyloid fibrils. Incubation of PFOs with phospholipids vesicles results in a loss of PFO immunoreactivity with a corresponding increase in alphaAPF immunoreactivity, suggesting that lipid vesicles catalyze the conversion of PFOs into APFs. The annular anti-protofibril antibody also recognizes heptameric alpha-hemolysin pores, but not monomers, suggesting that the antibody recognizes an epitope that is specific for a beta barrel structural motif.
Figures
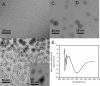
FIGURE 1.
Electron micrograph of PFOs and APFs and circular dichroism spectrum of APFs. A, Aβ42 PFO preparation used as the starting material for APF preparation. B, Aβ42 APFs prepared by mixing with 5% hexanes appear as ring or pore-like structures of varying diameter from 8 to 25 nm. They are obtained in a relatively homogenous and pure population, although small amounts of 3–5-nm spherical oligomers are also observed in the population. Immediately after preparation, the APFs have a rough or beaded appearance that is 3–5 nm in diameter that suggests that APFs form by the coalescence of prefibrillar spherical oligomers as previously suggested. Inset, after incubation for 1 week at 25 °C, the APFs lose the rough, beaded appearance and become smooth. Figures that appear to result from the concatenation of multiple APFs are also observed. C, Aβ40 APFs after 1 week of incubation. D, α-synuclein APFs after 1 week of incubation. E, circular dichroism spectrum of APFs demonstrating the β-sheet character of APFs.
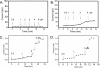
FIGURE 2.
Comparison of the effect of APFs and PFOs on the conductivity of black lipid membranes. A, Aβ42 APFs. No significant change in conductivity was observed. B, α-synuclein annular protofibrils. A slight increase in conductivity was observed. C, Aβ42 PFOs.D, α-synuclein PFOs. Arrows indicate the time of addition of increasing amounts of APFs or PFOs at the final concentrations indicated. PFOs increase membrane conductivity in a concentration-dependent fashion.
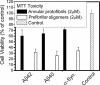
FIGURE 3.
Comparison of the toxicity of APFs and PFOs. Cytotoxicity was measured using SHSY5Y cells and the MTT assay as described under “Experimental Procedures” using 2 μ
m
APFs or PFOs. PFOs are significantly more toxic than APFs (p < 0.05).
FIGURE 4.
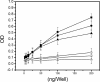
Specificity of anti-annular protofibril antisera. The immunoreactivity of the anti-annular protofibril antisera (anti-APF) was characterized by ELISA assay as described under “Experimental Procedures.” Anti-APF antisera reacts selectively with APFs (▪). Weak activity was observed with PFOs (▴). No reactivity is observed with monomers (♦) or fibrils (▾).
FIGURE 5.
αAPF antiserum recognizes conformation-dependent generic epitopes that are independent of specific amino acid sequences. APFs and monomeric proteins and peptides were analyzed by ELISA for Aβ40 (▪), islet amyloid polypeptide (•), and a-synuclein (▴). Closed symbols, APFs. Open symbols, monomeric proteins and peptides.
FIGURE 6.
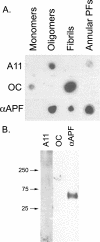
αAPF recognizes an epitope specifically associated with APFs. A, dot blot analysis of Aβ42 samples of monomer, PFOs, fibrils, and APFs with A11, OC, and αAPF. APFs are not stained by OC and only weakly stain with A11. B, Western blot analysis of Aβ42 APFs with A11, OC, and αAPF antibodies. αAPF specifically stains a band at ∼60 kDa that is not recognized by A11 or OC.
FIGURE 7.
αAPF fails to stain amyloid plaque deposits in AD brain. 6E10 stains both compact and diffuse amyloid plaque deposits (left panel), whereas no specific staining of amyloid deposits was observed for αAPF (right panel). Bar = 20 μm.
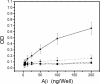
FIGURE 8.
Phosphatidylcholine vesicles catalyze the conformational conversion of PFOs to APFs. A sample of 66 μ
m
Aβ42 PFOs was mixed with 10 volumes of a solution of 25 m
m
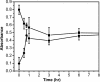
phosphatidylcholine liposomes, and samples were taken at different times and analyzed by ELISA with A11 (▪) or αAPF (•) antibodies. The loss of A11 immunoreactivity is correlated with an increase in αAPF reactivity.
FIGURE 9.
αAPF stains heptameric α-hemolysin pores. A, αAPF stains the mature pore band that runs at ∼250 kDa and only weakly stains a partially misfolded protomer band at 48 kDa. In contrast, A11 weakly stains the mature pore and intensely stains the protomer bands. Neither antibody stains the 33-kDa monomer band which is the major species stained by Coomassie Blue. B, incubation of α-hemolysin with deoxycholate promotes the 250-kDa heptameric pores stained by αAPF in a time-dependent fashion.
FIGURE 10.
Schematic diagram of annular protofibril formation at the surface of membranes. PFOs interact with the membrane, which induces a conformation change giving rise to the αAPF epitope. Additional PFOs are recruited to the membrane surface. When a sufficient number of oligomers have adsorbed, the aggregates form a pore-like structure.
Similar articles
- Fibrillar oligomers nucleate the oligomerization of monomeric amyloid beta but do not seed fibril formation.
Wu JW, Breydo L, Isas JM, Lee J, Kuznetsov YG, Langen R, Glabe C. Wu JW, et al. J Biol Chem. 2010 Feb 26;285(9):6071-9. doi: 10.1074/jbc.M109.069542. Epub 2009 Dec 15. J Biol Chem. 2010. PMID: 20018889 Free PMC article. - Structural classification of toxic amyloid oligomers.
Glabe CG. Glabe CG. J Biol Chem. 2008 Oct 31;283(44):29639-43. doi: 10.1074/jbc.R800016200. Epub 2008 Aug 22. J Biol Chem. 2008. PMID: 18723507 Free PMC article. Review. - Permeabilization of lipid bilayers is a common conformation-dependent activity of soluble amyloid oligomers in protein misfolding diseases.
Kayed R, Sokolov Y, Edmonds B, McIntire TM, Milton SC, Hall JE, Glabe CG. Kayed R, et al. J Biol Chem. 2004 Nov 5;279(45):46363-6. doi: 10.1074/jbc.C400260200. Epub 2004 Sep 21. J Biol Chem. 2004. PMID: 15385542 - Astrocytes contain amyloid-β annular protofibrils in Alzheimer's disease brains.
Lasagna-Reeves CA, Kayed R. Lasagna-Reeves CA, et al. FEBS Lett. 2011 Oct 3;585(19):3052-7. doi: 10.1016/j.febslet.2011.08.027. Epub 2011 Aug 24. FEBS Lett. 2011. PMID: 21872592 - New Mechanism of Amyloid Fibril Formation.
Galzitskaya O. Galzitskaya O. Curr Protein Pept Sci. 2019;20(6):630-640. doi: 10.2174/1389203720666190125160937. Curr Protein Pept Sci. 2019. PMID: 30686252 Review.
Cited by
- Thermodynamically stable amyloid-β monomers have much lower membrane affinity than the small oligomers.
Sarkar B, Das AK, Maiti S. Sarkar B, et al. Front Physiol. 2013 Apr 18;4:84. doi: 10.3389/fphys.2013.00084. eCollection 2013. Front Physiol. 2013. PMID: 23781202 Free PMC article. - Brain amyloid-β oligomers in ageing and Alzheimer's disease.
Lesné SE, Sherman MA, Grant M, Kuskowski M, Schneider JA, Bennett DA, Ashe KH. Lesné SE, et al. Brain. 2013 May;136(Pt 5):1383-98. doi: 10.1093/brain/awt062. Epub 2013 Apr 9. Brain. 2013. PMID: 23576130 Free PMC article. - Spontaneous self-assembly of amyloid β (1-40) into dimers.
Hashemi M, Zhang Y, Lv Z, Lyubchenko YL. Hashemi M, et al. Nanoscale Adv. 2019 Sep 17;1(10):3892-3899. doi: 10.1039/c9na00380k. eCollection 2019 Oct 9. Nanoscale Adv. 2019. PMID: 36132110 Free PMC article. - A causative link between the structure of aberrant protein oligomers and their toxicity.
Campioni S, Mannini B, Zampagni M, Pensalfini A, Parrini C, Evangelisti E, Relini A, Stefani M, Dobson CM, Cecchi C, Chiti F. Campioni S, et al. Nat Chem Biol. 2010 Feb;6(2):140-7. doi: 10.1038/nchembio.283. Epub 2010 Jan 10. Nat Chem Biol. 2010. PMID: 20081829 - General Principles Underpinning Amyloid Structure.
Taylor AIP, Staniforth RA. Taylor AIP, et al. Front Neurosci. 2022 Jun 2;16:878869. doi: 10.3389/fnins.2022.878869. eCollection 2022. Front Neurosci. 2022. PMID: 35720732 Free PMC article. Review.
References
- Dobson, C. M. (2006) Protein Pept. Lett. 13 219–227 - PubMed
- Hardy, J. (2005) Biochem. Soc. Trans. 33 578–581 - PubMed
- Chiti, F., Stefani, M., Taddei, N., Ramponi, G., and Dobson, C. M. (2003) Nature 424 805–808 - PubMed
- Terry, R. D. (1996) J. Neuropathol. Exp. Neurol. 55 1023–1025 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources