A stress-responsive RNA switch regulates VEGFA expression - PubMed (original) (raw)
. 2009 Feb 12;457(7231):915-9.
doi: 10.1038/nature07598. Epub 2008 Dec 21.
Affiliations
- PMID: 19098893
- PMCID: PMC2858559
- DOI: 10.1038/nature07598
A stress-responsive RNA switch regulates VEGFA expression
Partho Sarothi Ray et al. Nature. 2009.
Abstract
Ligand binding to structural elements in the non-coding regions of messenger RNA modulates gene expression. Ligands such as free metabolites or other small molecules directly bind and induce conformational changes in regulatory RNA elements known as riboswitches. Other types of RNA switches are activated by complexed metabolites-for example, RNA-ligated metabolites such as aminoacyl-charged transfer RNA in the T-box system, or protein-bound metabolites in the glucose- or amino-acid-stimulated terminator-anti-terminator systems. All of these switch types are found in bacteria, fungi and plants. Here we report an RNA switch in human vascular endothelial growth factor-A (VEGFA, also known as VEGF) mRNA 3' untranslated region (UTR) that integrates signals from interferon (IFN)-gamma and hypoxia to regulate VEGFA translation in myeloid cells. Analogous to riboswitches, the VEGFA 3' UTR undergoes a binary conformational change in response to environmental signals. However, the VEGFA 3' UTR switch is metabolite-independent, and the conformational change is dictated by mutually exclusive, stimulus-dependent binding of proteins, namely, the IFN-gamma-activated inhibitor of translation complex and heterogeneous nuclear ribonucleoprotein L (HNRNPL, also known as hnRNP L). We speculate that the VEGFA switch represents the founding member of a family of signal-mediated, protein-dependent RNA switches that evolved to regulate gene expression in multicellular animals in which the precise integration of disparate inputs may be more important than the rapidity of response.
Figures
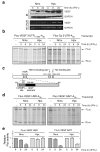
Figure 1. Suppression of GAIT-mediated translation silencing of VEGF by hypoxia
a, VEGF in lysates from U937 cells treated with IFN-γ in normoxia (Nmx.) or hypoxia (Hpx.) was determined by immunoblot (IB) and RT-PCR; GAPDH was probed as control. b, In vitro translation of Fluc reporter RNAs bearing VEGF 3’UTR11-900 (left) or Cp 3’UTR (right) in presence of cytosolic lysates and control Rluc RNA lacking a 3’UTR. c, Schematic of RNA elements in VEGF 3’UTR and HSR. d, In vitro translation of reporter RNAs bearing VEGF HSR (left) or GAIT element (right) in presence of cytosolic lysates. e, U937 cells were nucleofected with pcDNA3-Fluc reporters bearing VEGF HSR (top) or GAIT element (bottom). Cells were co-transfected with a plasmid expressing Rluc under SV40 promoter. Relative luciferase activity (Fluc/Rluc) was expressed as mean ± s.d. (3 experiments).
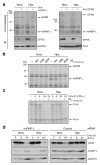
Figure 2. hnRNP L binding to HSR restores VEGF translation in hypoxia
a, Lysates from IFN-γ-treated cells were subjected to UV-crosslinking with [32P]UTP-labeled VEGF HSR RNA before and after immunodepletion with anti-hnRNP L (left) or anti-EPRS (right) antibodies. Effective depletion was shown by IB. b, Excess DNA oligomer antisense to hnRNP L binding site (AS3’UTR,332-357) blocks binding of hnRNP L to HSR RNA. c, Inhibition of hnRNP L binding by AS3’UTR,332-357 restores translational silencing of reporter RNA in hypoxia. d, siRNA-mediated knockdown of hnRNP L induces translational repression of VEGF in hypoxic cells. Lysates from cells transfected with hnRNP L (left) and control (right) siRNAs were immunoblotted with anti-VEGF, -hnRNP L and -GAPDH antibodies.
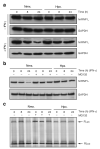
Figure 3. hnRNP L is regulated by stimulus-dependent proteasomal degradation
a, Lysates from U937 cells incubated in the absence or presence of IFN-γ were immunoblotted with anti-hnRNP L and -GAPDH antibodies. b, Immunoblot of lysates from cells treated with MG132 (200 nM). c, In vitro translation of VEGF HSR reporter RNA in presence of cell lysates.
Figure 4. Protein-dependent switching of the VEGF 3’UTR HSR
a, Secondary structure of VEGF HSR predicted by Mfold shows GAIT element (green), hnRNP L binding site (red), and stem stability sequence (blue). TP is lowest free energy conformer predicted by Mfold (left). TS conformer was generated by introducing experimentally-determined base-pairing constraints in GAIT element stem (right). Strong and weak RNase cleavage sites are marked by red and blue circles, respectively. Key signature cleavage sites are indicated (*, **). b, 32P-endlabeled VEGF HSR RNA was probed with RNase A under non-denaturing (lane 2) and denaturing (lane 3) conditions. Cleavages corresponding to predicted signature sites are indicated (*, **). c, RNase A probing of VEGF HSR RNA under non-denaturing conditions, or after the RNA was denatured and renatured. d, VEGF HSR RNA was incubated with cell lysates treated with IFN-γ for 24 h under normoxia or hypoxia, or with lysates immunodepleted with anti-hnRNP L and anti-EPRS antibodies, and subjected to RNase A-mediated cleavage under non-denaturing conditions after protein removal. e, Proposed pathway that switches the VEGF HSR to the TP conformer in the presence of IFN-γ and hypoxia (left), or to the TS conformer in the presence of IFN-γ and normoxia (right). f, Truth table showing AND NOT Boolean logic function of the VEGF RNA switch integrating signals from IFN-γ and hypoxia.
Similar articles
- The HILDA complex coordinates a conditional switch in the 3'-untranslated region of the VEGFA mRNA.
Yao P, Potdar AA, Ray PS, Eswarappa SM, Flagg AC, Willard B, Fox PL. Yao P, et al. PLoS Biol. 2013;11(8):e1001635. doi: 10.1371/journal.pbio.1001635. Epub 2013 Aug 20. PLoS Biol. 2013. PMID: 23976881 Free PMC article. - hnRNP L-mediated RNA switches function as a hypoxia-induced translational regulon.
Venkata Subbaiah KC, Wu J, Potdar A, Yao P. Venkata Subbaiah KC, et al. Biochem Biophys Res Commun. 2019 Aug 27;516(3):753-759. doi: 10.1016/j.bbrc.2019.06.106. Epub 2019 Jun 26. Biochem Biophys Res Commun. 2019. PMID: 31255281 Free PMC article. - Interplay between miR-574-3p and hnRNP L regulates VEGFA mRNA translation and tumorigenesis.
Yao P, Wu J, Lindner D, Fox PL. Yao P, et al. Nucleic Acids Res. 2017 Jul 27;45(13):7950-7964. doi: 10.1093/nar/gkx440. Nucleic Acids Res. 2017. PMID: 28520992 Free PMC article. - The GAIT translational control system.
Arif A, Yao P, Terenzi F, Jia J, Ray PS, Fox PL. Arif A, et al. Wiley Interdiscip Rev RNA. 2018 Mar;9(2):e1441. doi: 10.1002/wrna.1441. Epub 2017 Nov 20. Wiley Interdiscip Rev RNA. 2018. PMID: 29152905 Free PMC article. Review. - A switch in time: detailing the life of a riboswitch.
Garst AD, Batey RT. Garst AD, et al. Biochim Biophys Acta. 2009 Sep-Oct;1789(9-10):584-91. doi: 10.1016/j.bbagrm.2009.06.004. Epub 2009 Jul 9. Biochim Biophys Acta. 2009. PMID: 19595806 Free PMC article. Review.
Cited by
- A decade of riboswitches.
Serganov A, Nudler E. Serganov A, et al. Cell. 2013 Jan 17;152(1-2):17-24. doi: 10.1016/j.cell.2012.12.024. Cell. 2013. PMID: 23332744 Free PMC article. Review. - A systematic search for RNA structural switches across the human transcriptome.
Khoroshkin M, Asarnow D, Zhou S, Navickas A, Winters A, Goudreau J, Zhou SK, Yu J, Palka C, Fish L, Borah A, Yousefi K, Carpenter C, Ansel KM, Cheng Y, Gilbert LA, Goodarzi H. Khoroshkin M, et al. Nat Methods. 2024 Sep;21(9):1634-1645. doi: 10.1038/s41592-024-02335-1. Epub 2024 Jul 16. Nat Methods. 2024. PMID: 39014073 Free PMC article. - Role of hypoxia and vascular endothelial growth factors in lymphangiogenesis.
Morfoisse F, Renaud E, Hantelys F, Prats AC, Garmy-Susini B. Morfoisse F, et al. Mol Cell Oncol. 2014 Aug 13;1(1):e29907. doi: 10.4161/mco.29907. eCollection 2014. Mol Cell Oncol. 2014. PMID: 27308316 Free PMC article. Review. - New functions of aminoacyl-tRNA synthetases beyond translation.
Guo M, Yang XL, Schimmel P. Guo M, et al. Nat Rev Mol Cell Biol. 2010 Sep;11(9):668-74. doi: 10.1038/nrm2956. Epub 2010 Aug 11. Nat Rev Mol Cell Biol. 2010. PMID: 20700144 Free PMC article. Review. - Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: Focus on hnRNP E1's multifunctional regulatory roles.
Chaudhury A, Chander P, Howe PH. Chaudhury A, et al. RNA. 2010 Aug;16(8):1449-62. doi: 10.1261/rna.2254110. Epub 2010 Jun 28. RNA. 2010. PMID: 20584894 Free PMC article. Review.
References
- Mandal M, Breaker RR. Gene regulation by riboswitches. Nat Rev Mol Cell Biol. 2004;5:451–463. - PubMed
- Grundy FJ, Henkin TM. Regulation of gene expression by effectors that bind to RNA. Curr Opin Microbiol. 2004;7:126–131. - PubMed
- Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. - PubMed
- Cromie MJ, Shi Y, Latifi T, Groisman EA. An RNA sensor for intracellular Mg2+ Cell. 2006;125:71–84. - PubMed
- Grundy FJ, Henkin TM. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993;74:475–482. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK053307/DK/NIDDK NIH HHS/United States
- R01 HL067725-04/HL/NHLBI NIH HHS/United States
- P01 HL076491/HL/NHLBI NIH HHS/United States
- P01 HL029582/HL/NHLBI NIH HHS/United States
- R01 HL67725/HL/NHLBI NIH HHS/United States
- R01 DK060596/DK/NIDDK NIH HHS/United States
- P01 HL29582/HL/NHLBI NIH HHS/United States
- P01 HL076491-050002/HL/NHLBI NIH HHS/United States
- R01 HL067725/HL/NHLBI NIH HHS/United States
- R01 DK60596/DK/NIDDK NIH HHS/United States
- R01 DK060596-08/DK/NIDDK NIH HHS/United States
- P01 HL029582-26A18735/HL/NHLBI NIH HHS/United States
- P01 HL76491/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources