Night/day changes in pineal expression of >600 genes: central role of adrenergic/cAMP signaling - PubMed (original) (raw)
. 2009 Mar 20;284(12):7606-22.
doi: 10.1074/jbc.M808394200. Epub 2008 Dec 22.
Steven L Coon, David A Carter, Ann Humphries, Jong-So Kim, Qiong Shi, Pascaline Gaildrat, Fabrice Morin, Surajit Ganguly, John B Hogenesch, Joan L Weller, Martin F Rath, Morten Møller, Ruben Baler, David Sugden, Zoila G Rangel, Peter J Munson, David C Klein
Affiliations
- PMID: 19103603
- PMCID: PMC2658055
- DOI: 10.1074/jbc.M808394200
Night/day changes in pineal expression of >600 genes: central role of adrenergic/cAMP signaling
Michael J Bailey et al. J Biol Chem. 2009.
Abstract
The pineal gland plays an essential role in vertebrate chronobiology by converting time into a hormonal signal, melatonin, which is always elevated at night. Here we have analyzed the rodent pineal transcriptome using Affymetrix GeneChip(R) technology to obtain a more complete description of pineal cell biology. The effort revealed that 604 genes (1,268 probe sets) with Entrez Gene identifiers are differentially expressed greater than 2-fold between midnight and mid-day (false discovery rate <0.20). Expression is greater at night in approximately 70%. These findings were supported by the results of radiochemical in situ hybridization histology and quantitative real time-PCR studies. We also found that the regulatory mechanism controlling the night/day changes in the expression of most genes involves norepinephrine-cyclic AMP signaling. Comparison of the pineal gene expression profile with that in other tissues identified 334 genes (496 probe sets) that are expressed greater than 8-fold higher in the pineal gland relative to other tissues. Of these genes, 17% are expressed at similar levels in the retina, consistent with a common evolutionary origin of these tissues. Functional categorization of the highly expressed and/or night/day differentially expressed genes identified clusters that are markers of specialized functions, including the immune/inflammation response, melatonin synthesis, photodetection, thyroid hormone signaling, and diverse aspects of cellular signaling and cell biology. These studies produce a paradigm shift in our understanding of the 24-h dynamics of the pineal gland from one focused on melatonin synthesis to one including many cellular processes.
Figures
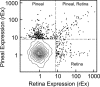
FIGURE 1.
Pineal rEx versus retina rEx. This figure demonstrates that a subset of genes is predominantly expressed in the pineal gland or retina or both. Data are based on results obtained in experiment C. The probe sets for the entire microarray are represented by 5% density contours (95% of probe sets are within the outermost contour line). Only symbols representing probe sets with rEx values greater than 8 for either the pineal gland or retina (∼1% of 31,099 probe sets on the Rat230_2 microarray) are shown; the remaining probe sets are not highly expressed in either tissue relative to other tissues. The plotted probe sets fall into three sectors as follows:Pineal, in which 218 genes (256 probe sets) are highly expressed primarily in the pineal gland; Pineal, Retina, in which 55 genes (63 probe sets) are highly expressed at similar levels in both the pineal gland and retina; and Retina, in which 93 genes (109 probe sets) are highly expressed primarily in the retina. The genes represented by these probe sets are listed in Table 4; supplemental Table S5 contains a detailed description of these genes and other unannotated probe sets. An interactive version of this figure that identifies each symbol is available at
sne.nichd.nih.gov
.
FIGURE 2.
Radiochemical in situ hybridization images. Each panel contains autoradiographs prepared from sections of rat brains through the pineal gland. The sections on the left are from animals killed during the day and those on the right are from animals killed during the night. The sections were incubated with antisense probes identified in the_bottom left-hand corner_ of the Day image. Probes are detailed in supplemental Table S1. The results of quantitation of the signal strength of the pineal labeling appear in Table 5. For further details see “Experimental Procedures.” Hab, habenula;ic, inferior colliculus; mhn, medial habenular nucleus;Raphe, dorsal raphe nucleus; sc, superior colliculus. These figures are available in high resolution at
sne.nichd.nih.gov
.
FIGURE 3.
Staining of pinealocytes and endothelial cells in the pineal gland. A, image presenting Aanat-labeling of pinealocytes. Dark field photomicrograph of radiochemical in situ hybridization of a part of the superficial pineal gland of the rat with an antisense probe binding to mRNA encoding AANAT. The section was dipped in a photographic emulsion and developed after exposure; the grains are seen using dark field visualization as white dots in the emulsion. Dense labeling is seen above the pineal parenchyma with no labeling of the perivascular space (per vasc). B, same section shown in A in transmitted light.Arrows indicate the perivascular space, which is not labeled in_A_. Cresyl violet counterstaining is used. C, labeling of the perivascular spaces; endothelial labeling image. Dark field photomicrograph of radiochemical in situ hybridization of the superficial pineal with an antisense probe binding to mRNA encoding Esm1. Note the labeling of the perivascular spaces (arrows). D, high power photomicrograph taken in transmitted light of part of the superficial pineal showing_Esm1_ labeling of the perivascular cells and only few grains above the pinealocytes. Cresyl violet counterstaining. Bars, A, B and_D_ = 20 μm. C = 50 μm. The probes used are described in supplemental Table S1.
FIGURE 4.
qRT-PCR analysis of transcripts that are night/day differentially expressed or have high rEx values or both. The lighting cycle is represented at the bottom of each column. Transcripts are identified by gene symbol. Each value is the mean ± S.E. of three determinations. Values were normalized to Actb, Gapdh, Hrpt1, and Rnr1. A single asterisk identifies statistically significant rhythmic patterns of gene expression (p < 0.01) based on log transformed raw values analyzed by one-way analysis of variance in JMP. For technical details, see “Experimental Procedures” and the supplemental material.
Similar articles
- Thyroid hormone and adrenergic signaling interact to control pineal expression of the dopamine receptor D4 gene (Drd4).
Kim JS, Bailey MJ, Weller JL, Sugden D, Rath MF, Møller M, Klein DC. Kim JS, et al. Mol Cell Endocrinol. 2010 Jan 15;314(1):128-35. doi: 10.1016/j.mce.2009.05.013. Epub 2009 May 29. Mol Cell Endocrinol. 2010. PMID: 19482058 Free PMC article. - Homeobox genes and melatonin synthesis: regulatory roles of the cone-rod homeobox transcription factor in the rodent pineal gland.
Rohde K, Møller M, Rath MF. Rohde K, et al. Biomed Res Int. 2014;2014:946075. doi: 10.1155/2014/946075. Epub 2014 Apr 30. Biomed Res Int. 2014. PMID: 24877149 Free PMC article. Review. - NGFI-B (Nurr77/Nr4a1) orphan nuclear receptor in rat pinealocytes: circadian expression involves an adrenergic-cyclic AMP mechanism.
Humphries A, Weller J, Klein D, Baler R, Carter DA. Humphries A, et al. J Neurochem. 2004 Nov;91(4):946-55. doi: 10.1111/j.1471-4159.2004.02777.x. J Neurochem. 2004. PMID: 15525348 - The Lhx4 homeobox transcript in the rat pineal gland: Adrenergic regulation and impact on transcripts encoding melatonin-synthesizing enzymes.
Hertz H, Carstensen MB, Bering T, Rohde K, Møller M, Granau AM, Coon SL, Klein DC, Rath MF. Hertz H, et al. J Pineal Res. 2020 Jan;68(1):e12616. doi: 10.1111/jpi.12616. Epub 2019 Nov 11. J Pineal Res. 2020. PMID: 31609018 Free PMC article. - Control of CREB phosphorylation and its role for induction of melatonin synthesis in rat pinealocytes.
Maronde E, Schomerus C, Stehle JH, Korf HW. Maronde E, et al. Biol Cell. 1997 Nov;89(8):505-11. doi: 10.1016/s0248-4900(98)80006-3. Biol Cell. 1997. PMID: 9618900 Review.
Cited by
- Microarray analysis of verbenalin-treated human amniotic epithelial cells reveals therapeutic potential for Alzheimer's Disease.
Ferdousi F, Kondo S, Sasaki K, Uchida Y, Ohkohchi N, Zheng YW, Isoda H. Ferdousi F, et al. Aging (Albany NY). 2020 Mar 29;12(6):5516-5538. doi: 10.18632/aging.102985. Epub 2020 Mar 29. Aging (Albany NY). 2020. PMID: 32224504 Free PMC article. - Regulation of diurnal energy balance by mitokines.
Klaus S, Igual Gil C, Ost M. Klaus S, et al. Cell Mol Life Sci. 2021 Apr;78(7):3369-3384. doi: 10.1007/s00018-020-03748-9. Epub 2021 Jan 19. Cell Mol Life Sci. 2021. PMID: 33464381 Free PMC article. Review. - Melatonin Uptake by Cells: An Answer to Its Relationship with Glucose?
Mayo JC, Aguado A, Cernuda-Cernuda R, Álvarez-Artime A, Cepas V, Quirós-González I, Hevia D, Sáinz RM. Mayo JC, et al. Molecules. 2018 Aug 10;23(8):1999. doi: 10.3390/molecules23081999. Molecules. 2018. PMID: 30103453 Free PMC article. Review. - Drastic neofunctionalization associated with evolution of the timezyme AANAT 500 Mya.
Falcón J, Coon SL, Besseau L, Cazaméa-Catalan D, Fuentès M, Magnanou E, Paulin CH, Boeuf G, Sauzet S, Jørgensen EH, Mazan S, Wolf YI, Koonin EV, Steinbach PJ, Hyodo S, Klein DC. Falcón J, et al. Proc Natl Acad Sci U S A. 2014 Jan 7;111(1):314-9. doi: 10.1073/pnas.1312634110. Epub 2013 Dec 18. Proc Natl Acad Sci U S A. 2014. PMID: 24351931 Free PMC article. - Circadian Plasticity in the Brain of Insects and Rodents.
Krzeptowski W, Hess G, Pyza E. Krzeptowski W, et al. Front Neural Circuits. 2018 May 2;12:32. doi: 10.3389/fncir.2018.00032. eCollection 2018. Front Neural Circuits. 2018. PMID: 29770112 Free PMC article. Review.
References
- Maronde, E., and Stehle, J. H. (2007) Trends Endocrinol. Metab. 18 142–149 - PubMed
- Arendt, J. (1994) Melatonin and the Mammalian Pineal Gland, 1st Ed., pp. 1–331, Chapman & Hall, New York
- Lincoln, G. A. (2006) Chronobiol. Int. 23 301–306 - PubMed
- Klein, D. C. (1985) CIBA Found. Symp. 117 38–56 - PubMed
- Moller, M., and Baeres, F. M. (2002) Cell Tissue Res. 309 139–150 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases