Regulation of SMN protein stability - PubMed (original) (raw)
Regulation of SMN protein stability
Barrington G Burnett et al. Mol Cell Biol. 2009 Mar.
Abstract
Spinal muscular atrophy (SMA) is caused by mutations of the survival of motor neuron (SMN1) gene and deficiency of full-length SMN protein (FL-SMN). All SMA patients retain one or more copies of the SMN2 gene, but the principal protein product of SMN2 lacks exon 7 (SMNDelta7) and is unable to compensate for a deficiency of FL-SMN. SMN is known to oligomerize and form a multimeric protein complex; however, the mechanisms regulating stability and degradation of FL-SMN and SMNDelta7 proteins have been largely unexplored. Using pulse-chase analysis, we characterized SMN protein turnover and confirmed that SMN was ubiquitinated and degraded by the ubiquitin proteasome system (UPS). The SMNDelta7 protein had a twofold shorter half-life than FL-SMN in cells despite similar intrinsic rates of turnover by the UPS in a cell-free assay. Mutations that inhibited SMN oligomerization and complex formation reduced the FL-SMN half-life. Furthermore, recruitment of SMN into large macromolecular complexes as well as increased association with several Gemin proteins was regulated in part by protein kinase A. Together, our data indicate that SMN protein stability is modulated by complex formation. Promotion of the SMN complex formation may be an important novel therapeutic strategy for SMA.
Figures
FIG. 1.
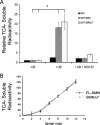
SMN is ubiquitinated and degraded by the UPS. (A) Treatment of 3813 SMA patient-derived fibroblast with the proteasome inhibitor, MG132 (10 μM); the cell-permeable calpain inhibitor, Calpeptin; the lysosome inhibitor, ammonium chloride (NH4Cl); and an autophagy inhibitor, 3-MA. Quantification is shown in lower panel. (B) 3813 cells treated with 1, 5, or 10 μM concentrations of the proteasome inhibitor lactacystin. The data represent mean ± the standard error of the mean (SEM) of five independent experiments. *, P < 0.05. (C) Cell lysates were denatured with 1% SDS and then renatured with 4.5% Triton X-100 prior to immunoprecipitation with anti-SMN antibody. In addition, cell lysis buffer was supplemented with 2.5 μM ubiquitin aldehyde to inhibit deubiquitinating enzymes. Western bots were probed with antibodies to SMN, Gemin 2, Gemin 3, and Gemin 6. (D) Western blot probed with an antibody to ubiquitin after immunoprecipitation of SMN from cells in the presence or absence of 10 μM lactacystin.
FIG. 2.
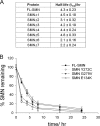
FL-SMN and SMNΔ7 are ubiquitinated and degraded by the proteasome. (A) Quantification of TCA-soluble SMN fragments after incubation of ubiquitinated or nonubiquitinated 125I-GST-FL-SMN with 5 nM 26S proteasome after 30 min. The data represent mean ± the SEM of three independent experiments. *, P < 0.05. (B) Rate of degradation of ubiquitinated 125I-GST-SMN and 125I-GST-SMNΔ7 after incubation with 5 nM purified 26S proteasome and precipitation, at the designated times, on ice with 10% TCA. The data represent mean ± the SEM of three independent experiments.
FIG. 3.
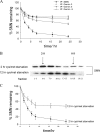
SMNΔ7 is more rapidly degraded than FL-SMN. (A) Pulse-chase analysis of endogenous SMN, myc-SMN, and GFP-SMN in HEK293T cells. (B) Pulse-chase analysis of myc-SMN and SMNΔ7. (C) Pulse-chase analysis of myc-SMN and SMNΔ7 in the absence or presence of 10 μM MG132. (D) Pulse-chase analysis of SMNΔ7 in HEK293T cells after knockdown of endogenous SMN 72 h after transfection of shRNA targeting SMN. The data represent mean ± the SEM of three independent experiments. Inset shows Western blots of endogenous SMN, the shRNA-resistant myc-SMNΔ7 (designated by an asterisk) and actin at various time points after SMN shRNA transfection.
FIG. 4.
Residues that are critical for oligomerization modulate SMN stabilization. (A) Pulse-chase analysis of SMN deletion mutants lacking exons 1 through 7. The data represent mean ± the SEM of three experiments. (B) Pulse-chase assay performed on FL-SMN, as well as SMN mutants Y272C and G279V, which show deficits in SMN complex formation, and E134K, which does not affect complex formation. The data represent mean ± the SEM of three experiments.
FIG. 5.
SMN is stabilized by incorporation into the SMN complex. (A) Pulse-chase assay performed using Gemin 2 and Gemin 3 antibodies to identify SMN that may be in complex. The data represent the means ± the SEM of four experiments. (B) SMN from cells starved of cysteine-methionine for 2 and 12 h and pulse-labeled with radioactive cysteine-methionine, followed by 10 to 30% sucrose gradient separation. (C) Pulse-chase analysis of SMN from cells starved of cysteine-methionine for 2 and 12 h. The data represent mean ± the SEM of five experiments.
FIG. 6.
SMN is stabilized by PKA. (A) Western blot of SMN from 3813 cells treated with forskolin (10 μM) for the indicated times. (B) Quantification of blot in panel A normalized to actin. (C) Western blot of SMN from cells overexpressing constitutively active PKA. (D) Autoradiograph of SMN and point mutants following in vitro PKA assay using purified catalytic subunit of PKA and purified GST-SMN as the substrate. (E) SMN turnover in the presence or absence of constitutively active PKA and the PKA inhibitor PKI.
FIG. 7.
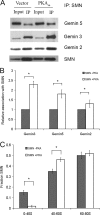
PKA increases the amount of SMN in complex. (A) Western blot of SMN immunoprecipitated from cells either expressing constitutively active PKA or the empty vector. Blots were probed with antibodies to Gemin 5, Gemin 3, Gemin 2, and SMN. (B) The relative association of gemins with SMN was obtained by normalizing the amount of coimmunoprecipitated Gemin to total SMN protein. Bar graph represents the relative amounts of coimmunoprecipitated Gemin in the presence of PKA to that in the absence of PKA. The data represent mean ± the SEM of three experiments. *, P < 0.05. (C) Quantification of SMN in sucrose gradient fractions from cells expressing constitutively active PKA or empty vector. The data represent mean ± the SEM of four experiments. *, P < 0.05.
Similar articles
- SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN.
Le TT, Pham LT, Butchbach ME, Zhang HL, Monani UR, Coovert DD, Gavrilina TO, Xing L, Bassell GJ, Burghes AH. Le TT, et al. Hum Mol Genet. 2005 Mar 15;14(6):845-57. doi: 10.1093/hmg/ddi078. Epub 2005 Feb 9. Hum Mol Genet. 2005. PMID: 15703193 - Ubiquitination Insight from Spinal Muscular Atrophy-From Pathogenesis to Therapy: A Muscle Perspective.
Bolado-Carrancio A, Tapia O, Rodríguez-Rey JC. Bolado-Carrancio A, et al. Int J Mol Sci. 2024 Aug 13;25(16):8800. doi: 10.3390/ijms25168800. Int J Mol Sci. 2024. PMID: 39201486 Free PMC article. Review. - Different Stability and Proteasome-Mediated Degradation Rate of SMN Protein Isoforms.
Locatelli D, Terao M, Kurosaki M, Zanellati MC, Pletto DR, Finardi A, Colciaghi F, Garattini E, Battaglia GS. Locatelli D, et al. PLoS One. 2015 Jul 27;10(7):e0134163. doi: 10.1371/journal.pone.0134163. eCollection 2015. PLoS One. 2015. PMID: 26214005 Free PMC article. - Ubiquitin-specific protease 9x deubiquitinates and stabilizes the spinal muscular atrophy protein-survival motor neuron.
Han KJ, Foster DG, Zhang NY, Kanisha K, Dzieciatkowska M, Sclafani RA, Hansen KC, Peng J, Liu CW. Han KJ, et al. J Biol Chem. 2012 Dec 21;287(52):43741-52. doi: 10.1074/jbc.M112.372318. Epub 2012 Oct 30. J Biol Chem. 2012. PMID: 23112048 Free PMC article. - Pathogenesis and therapeutic targets in spinal muscular atrophy (SMA).
Lefebvre S, Sarret C. Lefebvre S, et al. Arch Pediatr. 2020 Dec;27(7S):7S3-7S8. doi: 10.1016/S0929-693X(20)30269-4. Arch Pediatr. 2020. PMID: 33357595 Review.
Cited by
- Small molecule screen reveals regulation of survival motor neuron protein abundance by Ras proteins.
Letso RR, Bauer AJ, Lunn MR, Yang WS, Stockwell BR. Letso RR, et al. ACS Chem Biol. 2013 May 17;8(5):914-22. doi: 10.1021/cb300374h. Epub 2013 Mar 29. ACS Chem Biol. 2013. PMID: 23496866 Free PMC article. - Bap1/SMN axis in Dpp4+ skeletal muscle mesenchymal cells regulates the neuromuscular system.
Kim JH, Kang JS, Yoo K, Jeong J, Park I, Park JH, Rhee J, Jeon S, Jo YW, Hann SH, Seo M, Moon S, Um SJ, Seong RH, Kong YY. Kim JH, et al. JCI Insight. 2022 May 23;7(10):e158380. doi: 10.1172/jci.insight.158380. JCI Insight. 2022. PMID: 35603786 Free PMC article. - Ddx20, an Olig2 binding factor, governs the survival of neural and oligodendrocyte progenitor cells via proper Mdm2 splicing and p53 suppression.
Bizen N, Bepari AK, Zhou L, Abe M, Sakimura K, Ono K, Takebayashi H. Bizen N, et al. Cell Death Differ. 2022 May;29(5):1028-1041. doi: 10.1038/s41418-021-00915-8. Epub 2022 Jan 1. Cell Death Differ. 2022. PMID: 34974536 Free PMC article. - Spinal muscular atrophy: an update on therapeutic progress.
Seo J, Howell MD, Singh NN, Singh RN. Seo J, et al. Biochim Biophys Acta. 2013 Dec;1832(12):2180-90. doi: 10.1016/j.bbadis.2013.08.005. Epub 2013 Aug 27. Biochim Biophys Acta. 2013. PMID: 23994186 Free PMC article. Review. - Assays for the identification and prioritization of drug candidates for spinal muscular atrophy.
Cherry JJ, Kobayashi DT, Lynes MM, Naryshkin NN, Tiziano FD, Zaworski PG, Rubin LL, Jarecki J. Cherry JJ, et al. Assay Drug Dev Technol. 2014 Aug;12(6):315-41. doi: 10.1089/adt.2014.587. Assay Drug Dev Technol. 2014. PMID: 25147906 Free PMC article. Review.
References
- Angelozzi, C., F. Borgo, F. D. Tiziano, A. Martella, G. Neri, and C. Brahe. 2008. Salbutamol increases SMN mRNA and protein levels in spinal muscular atrophy cells. J. Med. Genet. 4529-31. - PubMed
- Baccon, J., L. Pellizzoni, J. Rappsilber, M. Mann, and G. Dreyfuss. 2002. Identification and characterization of Gemin7, a novel component of the survival of motor neuron complex. J. Biol. Chem. 27731957-31962. - PubMed
- Blom, N., S. Gammeltoft, and S. Brunak. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 2941351-1362. - PubMed
- Chang, H. C., W. C. Hung, Y. J. Chuang, and Y. J. Jong. 2004. Degradation of survival motor neuron (SMN) protein is mediated via the ubiquitin/proteasome pathway. Neurochem. Int. 451107-1112. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources