Molecular mechanism for the umami taste synergism - PubMed (original) (raw)
Molecular mechanism for the umami taste synergism
Feng Zhang et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Umami is one of the 5 basic taste qualities. The umami taste of L-glutamate can be drastically enhanced by 5' ribonucleotides and the synergy is a hallmark of this taste quality. The umami taste receptor is a heteromeric complex of 2 class C G-protein-coupled receptors, T1R1 and T1R3. Here we elucidate the molecular mechanism of the synergy using chimeric T1R receptors, site-directed mutagenesis, and molecular modeling. We propose a cooperative ligand-binding model involving the Venus flytrap domain of T1R1, where L-glutamate binds close to the hinge region, and 5' ribonucleotides bind to an adjacent site close to the opening of the flytrap to further stabilize the closed conformation. This unique mechanism may apply to other class C G-protein-coupled receptors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
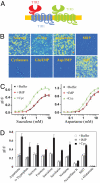
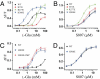
Fig. 1.
The sweet-umami chimeric receptor. (A) A schematic of sweet-umami chimeric receptor. Human T1R1 sequence is in blue, T1R2 is in red, and T1R3 is in green. (B) Responses of sweet-umami chimeric receptor to different sweet and umami stimuli. HEK293 cells stably expressing hT1R2–1/hT1R3 and G16gust25 were assayed for intracellular calcium increase in response to sucrose (200 mM), D-tryptophan (10 mM), aspartame (2.5 mM), S819 (25 μM), cyclamate (80 mM), L-glutamate/IMP (100/10 mM), L-aspartate/IMP (100/10 mM), and S807 (6 μM). (C) Dose-dependent responses of the hT1R2–1/hT1R3 stable cell line to sucralose and aspartame in the absence and presence of IMP (10 mM) and cyclamate (10 mM). The EC50s (mM) are 0.96, 0.97 (with IMP), and 0.41 (with cyclamate) for sucralose, and 2.5, 2.6 (with IMP), and 1.6 (with cyclamate) for aspartame. (D) Responses of the hT1R2–1/hT1R3 stable cell line to more sweet and umami taste stimuli in the absence and presence of IMP (10 mM) and cyclamate (10 mM). Concentrations: aspartame (2.5 mM), D-tryptophan (10 mM), sucrose (200 mM), fructose (200 mM), sucralose (1 mM), saccharin (2 mM), neotame (0.1 mM), acesulfame K (2.5 mM), S807 (3 μM), L-glutamate (100 mM). The values in C to D represent the mean ± SD of ΔF/F for 4 independent responses recorded using Fluorescence Imaging Plate Reader (FLIPR).
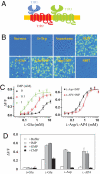
Fig. 2.
The umami-sweet chimeric receptor. (A) A schematic of umami-sweet chimeric receptor. (B) Responses of umami-sweet chimeric receptor to different sweet and umami stimuli. HEK293 cells stably expressing hT1R1–2/rT1R3 and G16gust44 were assayed for intracellular calcium increase in response to sucrose (200 mM), D-tryptophan (10 mM), aspartame (2.5 mM), S819 (25 μM), L-glutamate (100 mM), L-glutamate/IMP (100/10 mM), L-aspartate/IMP (100/10 mM), and S807 (6 μM). (C) Dose-dependent responses of the umami-sweet chimeric receptor. The EC50s (mM) for L-glutamate are 3.2, 0.9, and 0.2 in the presence of 0, 0.1 and 1 mM IMP respectively. The EC50s (mM) for L-AP4 and L-aspartate are 1.0 and 0.5 in the presence of 10 mM IMP. (D) Responses of hT1R1–2/rT1R3 stable cell line to umami taste stimuli in the absence and presence of 10 mM IMP or GMP. D-glutamate was used as a negative control for L-glutamate, and CMP was used as a negative control for IMP and GMP. The values in C to D represent the mean ± SD of ΔF/F for four independent responses recorded using FLIPR.
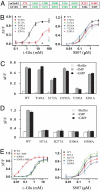
Fig. 3.
Mutagenesis analysis. (A) Critical ligand-binding residues in mGluR1 VFT domain and equivalent residues in human T1R1. mGluR1 residues that interact with the carboxylate side chain are in red, and those that interact with the α-amino acid moiety are in green. All 5 residues in green are conserved in T1R1. (B) The dose responses of mutant umami receptors that affect L-glutamate activity. (C) Mutant receptors that affect L-glutamate activity can still be enhanced by IMP and GMP. The L-glutamate concentrations used in the test are 0.4 (WT), 1.0 (T149A), and 100 mM (S172A, D192A, Y220A, and E301A). IMP and GMP concentration is 10 mM. (D) Human T1R1 mutations that affect the enhancement activity of IMP and GMP. The L-glutamate concentrations (mM) used in the test are 0.4 (WT), 10 (H71A), 3.0 (S306A), and 1.0 (R277A and H308A), roughly EC10 based on their respective dose-response curves. IMP and GMP concentration is 10 mM. (E) The dose responses of human T1R1 mutant receptors that affect the enhancement activity of IMP and GMP. The EC50s of WT receptor are 1.3 and 0.02 mM in the absence and presence of 1 mM IMP, respectively. The EC50s of H308A mutant receptor are 1.2 and 0.9 mM in the absence and presence of 1 mM IMP, respectively. All 4 mutants responded to S807 similarly as the wild type. The values in B to D represent the mean ± SD of ΔF/F for 4 independent responses recorded using FLIPR.
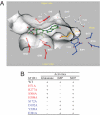
Fig. 4.
A molecular model of T1R1 VFT domain. (A) Key residues for L-glutamate (blue) and for IMP (red) binding. The VFT domain is oriented so that the opening to solvent is horizontal, with the upper lobe on the top and the lower lobe on the bottom. The flytrap hinge region is to the right, and the flytrap opening is to the left. L-glutamate (golden) is located deep inside the VFT domain near the hinge region, while IMP (green) is located close to the opening of the VFT domain. (B) A summary of the mutagenesis data of the key residues. Residues important for L-glutamate activity are in blue, while those for IMP are in red.
Fig. 5.
Mutations of pincer residues to reverse the charge. (A) The L-glutamate dose responses of 3 mutants with reversed charges. The EC50s are 2.6 (WT), 22 (H71E), 12 (R277E), and 0.8 mM (H308E). (B) The S807 dose responses of 3 mutants with reversed charges. The EC50s are 1.3 (WT), 1.1 (H71E), 0.6 (R277E), and 1.2 μM (H308E). (C) The L-glutamate dose responses of H308E in the absence and presence of 1 mM IMP. Gα15 cells were transiently transfected with human T1R1 WT or H308E mutant together with human T1R3. The EC50s are 4.3 and 4.0 mM for the mutant receptor in the absence and presence of IMP. (D) The S807 dose responses of the H308E mutant. Gα15 cells were transiently transfected with human T1R1 WT or H308E mutant together with human T1R3. The EC50s are 0.6 μM for WT and 0.5 μM for H308E mutant receptor. All values represent the mean ± SD of ΔF/F for 4 independent responses recorded using FLIPR.
Similar articles
- T1R receptors mediate mammalian sweet and umami taste.
Li X. Li X. Am J Clin Nutr. 2009 Sep;90(3):733S-737S. doi: 10.3945/ajcn.2009.27462G. Epub 2009 Aug 5. Am J Clin Nutr. 2009. PMID: 19656838 - Two distinct determinants of ligand specificity in T1R1/T1R3 (the umami taste receptor).
Toda Y, Nakagita T, Hayakawa T, Okada S, Narukawa M, Imai H, Ishimaru Y, Misaka T. Toda Y, et al. J Biol Chem. 2013 Dec 27;288(52):36863-77. doi: 10.1074/jbc.M113.494443. Epub 2013 Nov 8. J Biol Chem. 2013. PMID: 24214976 Free PMC article. - Molecular mechanism of the allosteric enhancement of the umami taste sensation.
Mouritsen OG, Khandelia H. Mouritsen OG, et al. FEBS J. 2012 Sep;279(17):3112-20. doi: 10.1111/j.1742-4658.2012.08690.x. Epub 2012 Jul 24. FEBS J. 2012. PMID: 22764741 - Multiple receptor systems for glutamate detection in the taste organ.
Yasuo T, Kusuhara Y, Yasumatsu K, Ninomiya Y. Yasuo T, et al. Biol Pharm Bull. 2008 Oct;31(10):1833-7. doi: 10.1248/bpb.31.1833. Biol Pharm Bull. 2008. PMID: 18827337 Review. - Molecular insights into human taste perception and umami tastants: A review.
Diepeveen J, Moerdijk-Poortvliet TCW, van der Leij FR. Diepeveen J, et al. J Food Sci. 2022 Apr;87(4):1449-1465. doi: 10.1111/1750-3841.16101. Epub 2022 Mar 17. J Food Sci. 2022. PMID: 35301715 Free PMC article. Review.
Cited by
- Umami the Fifth Basic Taste: History of Studies on Receptor Mechanisms and Role as a Food Flavor.
Kurihara K. Kurihara K. Biomed Res Int. 2015;2015:189402. doi: 10.1155/2015/189402. Epub 2015 Jul 12. Biomed Res Int. 2015. PMID: 26247011 Free PMC article. Review. - Chloride ions evoke taste sensations by binding to the extracellular ligand-binding domain of sweet/umami taste receptors.
Atsumi N, Yasumatsu K, Takashina Y, Ito C, Yasui N, Margolskee RF, Yamashita A. Atsumi N, et al. Elife. 2023 Feb 28;12:e84291. doi: 10.7554/eLife.84291. Elife. 2023. PMID: 36852482 Free PMC article. - Genome and proteome of the chlorophyll f-producing cyanobacterium Halomicronema hongdechloris: adaptative proteomic shifts under different light conditions.
Chen M, Hernandez-Prieto MA, Loughlin PC, Li Y, Willows RD. Chen M, et al. BMC Genomics. 2019 Mar 12;20(1):207. doi: 10.1186/s12864-019-5587-3. BMC Genomics. 2019. PMID: 30866821 Free PMC article. - Direct binding of calmodulin to the cytosolic C-terminal regions of sweet/umami taste receptors.
Yoshida A, Ito A, Yasui N, Yamashita A. Yoshida A, et al. J Biochem. 2023 Oct 31;174(5):451-459. doi: 10.1093/jb/mvad060. J Biochem. 2023. PMID: 37527916 Free PMC article. - Effects and Mechanisms of Tastants on the Gustatory-Salivary Reflex in Human Minor Salivary Glands.
Satoh-Kuriwada S, Shoji N, Miyake H, Watanabe C, Sasano T. Satoh-Kuriwada S, et al. Biomed Res Int. 2018 Jan 31;2018:3847075. doi: 10.1155/2018/3847075. eCollection 2018. Biomed Res Int. 2018. PMID: 29651428 Free PMC article. Clinical Trial.
References
- Yamaguchi S, Ninomiya K. Umami and food palatability. J Nutr. 2000;130:921S–926S. - PubMed
- Chandrashekar J, Hoon MA, Ryba NJ, Zuker CS. The receptors and cells for mammalian taste. Nature. 2006;444:288–294. - PubMed
- Nelson G, et al. An amino-acid taste receptor. Nature. 2002;416:199–202. - PubMed
- Nelson G, et al. Mammalian sweet taste receptors. Cell. 2001;106:381–390. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases