Population genetics of Trypanosoma brucei gambiense, the agent of sleeping sickness in Western Africa - PubMed (original) (raw)
Population genetics of Trypanosoma brucei gambiense, the agent of sleeping sickness in Western Africa
Mathurin Koffi et al. Proc Natl Acad Sci U S A. 2009.
Abstract
Human African trypanosomiasis, or sleeping sickness caused by Trypanosoma brucei gambiense, occurs in Western and Central Africa. T. brucei s.l. displays a huge diversity of adaptations and host specificities, and questions about its reproductive mode, dispersal abilities, and effective size remain under debate. We have investigated genetic variation at 8 microsatellite loci of T. b. gambiense strains isolated from human African trypanosomiasis patients in the Ivory Coast and Guinea, with the aim of knowing how genetic information was partitioned within and between individuals in both temporal and spatial scales. The results indicate that (i) migration of T. b. gambiense group 1 strains does not occur at the scale of West Africa, and that even at a finer scale (e.g., within Guinea) migration is restricted; (ii) effective population sizes of trypanosomes, as reflected by infected hosts, are probably higher than what the epidemiological surveys suggest; and (iii) T. b. gambiense group 1 is most likely a strictly clonally reproducing organism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
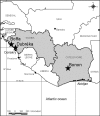
Fig. 1.
Localization of sampling areas (stars). (Drawing by Fabrice Courtin, Bobo-Dioulasso, Burkina Faso).
Fig. 2.
_F_IS per locus and over all 7 polymorphic loci (All) (Micbg6 and Trbpa1/2 excluded), averaged over the 6 subsamples. The residual variation across the 6 remaining loci is mainly explained (91%) by the corresponding genetic diversity (in clonal populations a positive relationship is indeed expected, see ref. 22). For each locus, 95% confidence intervals (CI) of the means are estimated with the jackknife method over the populations' standard error. Over all loci, CI was obtained by bootstrap over loci. Mean _F_IS and 95% bootstrap CI were also computed for Bonon 2000–2002 subsamples (Bonon) and compared to the _F_IS computed when GPS coordinates of patients are taken into account (i.e., grouping the most proximate isolates into smaller subunits) (Bonon GPS).
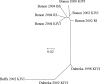
Fig. 3.
Unrooted NJTREE representation of genetic distance between the different subsamples of T. b. brucei in Guinea (Boffa and Dubreka) and in the Ivory Coast (Bonon) in different years (1998, 2002, and 2004) and with different sampling techniques (KIVI, RI, and BS) using Cavalli-Sforza and Edwards' (24) chord-distance matrix.
Fig. 4.
Effective population size (N e) obtained with the _F_IS-based method (see Materials and Methods Eq. 1) (“model”), with u = 10−3 and u = 10−4, and with Waples' method from temporally spaced samples (with MLG as a single locus), using trypanosome's life cycle as the generation time with the shortest (sgt = 37 days) or largest (lgt = 49 days) generation times (see text). Black squares are the means with 95% CIs (small lines) (averaged over 2000–2002, 2000–2004, and 2002–2004 for Bonon). The dotted line corresponds to the estimated number of infected persons in the different areas according to epidemiological surveys. For Waples' method, CI comes from a χ2 distribution with a degrees of freedom (a is the number of alleles, in this case of different MLG's) (25). For the _F_IS-based method, CIs correspond to those of _F_IS obtained by bootstrap over loci.
Similar articles
- Human and animal Trypanosomes in Côte d'Ivoire form a single breeding population.
Capewell P, Cooper A, Duffy CW, Tait A, Turner CM, Gibson W, Mehlitz D, Macleod A. Capewell P, et al. PLoS One. 2013 Jul 2;8(7):e67852. doi: 10.1371/journal.pone.0067852. Print 2013. PLoS One. 2013. PMID: 23844111 Free PMC article. - Genetic characterization of Trypanosoma brucei gambiense and clinical evolution of human African trypanosomiasis in Côte d'Ivoire.
Jamonneau V, Garcia A, Ravel S, Cuny G, Oury B, Solano P, N'Guessan P, N'Dri L, Sanon R, Frézil JL, Truc P. Jamonneau V, et al. Trop Med Int Health. 2002 Jul;7(7):610-21. doi: 10.1046/j.1365-3156.2002.00905.x. Trop Med Int Health. 2002. PMID: 12100445 - Free-ranging pigs identified as a multi-reservoir of Trypanosoma brucei and Trypanosoma congolense in the Vavoua area, a historical sleeping sickness focus of Côte d'Ivoire.
Traoré BM, Koffi M, N'Djetchi MK, Kaba D, Kaboré J, Ilboudo H, Ahouty BA, Koné M, Coulibaly B, Konan T, Segard A, Kouakou L, De Meeûs T, Ravel S, Solano P, Bart JM, Jamonneau V. Traoré BM, et al. PLoS Negl Trop Dis. 2021 Dec 22;15(12):e0010036. doi: 10.1371/journal.pntd.0010036. eCollection 2021 Dec. PLoS Negl Trop Dis. 2021. PMID: 34937054 Free PMC article. - Trypanosoma brucei gambiense Group 2: The Unusual Suspect.
Jamonneau V, Truc P, Grébaut P, Herder S, Ravel S, Solano P, De Meeus T. Jamonneau V, et al. Trends Parasitol. 2019 Dec;35(12):983-995. doi: 10.1016/j.pt.2019.09.002. Epub 2019 Oct 24. Trends Parasitol. 2019. PMID: 31668893 Review. - The challenge of Trypanosoma brucei gambiense sleeping sickness diagnosis outside Africa.
Lejon V, Boelaert M, Jannin J, Moore A, Büscher P. Lejon V, et al. Lancet Infect Dis. 2003 Dec;3(12):804-8. doi: 10.1016/s1473-3099(03)00834-x. Lancet Infect Dis. 2003. PMID: 14652206 Review.
Cited by
- Population genetic structure and temporal stability among Trypanosoma brucei rhodesiense isolates in Uganda.
Kato CD, Alibu VP, Nanteza A, Mugasa CM, Matovu E. Kato CD, et al. Parasit Vectors. 2016 May 3;9:259. doi: 10.1186/s13071-016-1542-1. Parasit Vectors. 2016. PMID: 27142001 Free PMC article. - Signatures of hybridization in Trypanosoma brucei.
Kay C, Peacock L, Williams TA, Gibson W. Kay C, et al. PLoS Pathog. 2022 Feb 9;18(2):e1010300. doi: 10.1371/journal.ppat.1010300. eCollection 2022 Feb. PLoS Pathog. 2022. PMID: 35139131 Free PMC article. - Phylogeography and taxonomy of Trypanosoma brucei.
Balmer O, Beadell JS, Gibson W, Caccone A. Balmer O, et al. PLoS Negl Trop Dis. 2011 Feb 8;5(2):e961. doi: 10.1371/journal.pntd.0000961. PLoS Negl Trop Dis. 2011. PMID: 21347445 Free PMC article. - Single-cell transcriptomics supports presence of cryptic species and reveals low levels of population genetic diversity in two testate amoebae morphospecies with large population sizes.
Weiner AKM, Sehein T, Cote-L'Heureux A, Sleith RS, Greco M, Malekshahi C, Ryan-Embry C, Ostriker N, Katz LA. Weiner AKM, et al. Evolution. 2023 Nov 2;77(11):2472-2483. doi: 10.1093/evolut/qpad158. Evolution. 2023. PMID: 37672006 Free PMC article. - Human and animal Trypanosomes in Côte d'Ivoire form a single breeding population.
Capewell P, Cooper A, Duffy CW, Tait A, Turner CM, Gibson W, Mehlitz D, Macleod A. Capewell P, et al. PLoS One. 2013 Jul 2;8(7):e67852. doi: 10.1371/journal.pone.0067852. Print 2013. PLoS One. 2013. PMID: 23844111 Free PMC article.
References
- Cattand P. L'épidémiologie de la trypanosomiase humaine africaine : Une histoire multifactorielle complexe. Médecine Tropicale. 2001;61:313–322. - PubMed
- Hoare CB. The Trypanosomes of Mammals: A zoological monograph. Oxford: Blackwell; 1972.
- Gibson WC. The significance of genetic exchange in trypanosomes. Parasitol Today. 1995;11:465–468.
- Gibson WC. Resolution of the species problem in African trypanosomes. Int J Parasitol. 2007;37:829–838. - PubMed
- Tibayrenc M. Population genetics of parasitic protozoa and other micro-organisms. Adv Parasitol. 1995;36:47–115. - PubMed