A conserved protonation-dependent switch controls drug binding in the Abl kinase - PubMed (original) (raw)
A conserved protonation-dependent switch controls drug binding in the Abl kinase
Yibing Shan et al. Proc Natl Acad Sci U S A. 2009.
Abstract
In many protein kinases, a characteristic conformational change (the "DFG flip") connects catalytically active and inactive conformations. Many kinase inhibitors--including the cancer drug imatinib--selectively target a specific DFG conformation, but the function and mechanism of the flip remain unclear. Using long molecular dynamics simulations of the Abl kinase, we visualized the DFG flip in atomic-level detail and formulated an energetic model predicting that protonation of the DFG aspartate controls the flip. Consistent with our model's predictions, we demonstrated experimentally that the kinetics of imatinib binding to Abl kinase have a pH dependence that disappears when the DFG aspartate is mutated. Our model suggests a possible explanation for the high degree of conservation of the DFG motif: that the flip, modulated by electrostatic changes inherent to the catalytic cycle, allows the kinase to access flexible conformations facilitating nucleotide binding and release.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
A proposed mechanism for the DFG flip based on crystal structures and our Abl simulations. Key structural features are highlighted, with salt bridges represented by red dotted lines. The ATP-binding site is located between the N-lobe and the C-lobe. The active structure (A) and the autoinhibited structure (C) differ in their DFG conformation, and interconvert via a DFG flip. Nevertheless, these structures both adopt conformations referred to as αC-in, in which helix αC is close to the ATP-binding site so that the Lys-271–Glu-286 salt bridge may form. (B) The structure shown is taken from the simulations, and is a proposed intermediate in the DFG flip. This structure differs from A and C in 2 significant ways. First, helix αC is positioned away from the ATP-binding site so that Glu-286 may form a salt bridge with Arg-386 (an αC-out conformation). Second, the outward displacement of helix αC creates a pocket at the base of the N-lobe, which we refer to as the N-pocket, and which in B is occupied by Phe-382. The PDB ID codes of the structures on which the figure is based are 2F4J (A) and 1OPK (C). (1OPK contains regulatory domains, but only the kinase domain is shown here.)
Fig. 2.
The DFG motif and the N-pocket. Both A and B show the base of the N-lobe, with the C-lobe removed to make the N-pocket and DFG motif fully visible (see Inset). (A) An αC-out conformation illustrating the N-pocket. In this conformation the close interactions between Phe-382 and Met-290 have been transiently broken, but Met-290 still presents a steric barrier to the entry of Phe-382 into the N-pocket. This structure was taken from simulation 1a after 250 ns. (B) Phe-382 occupying the N-pocket, as enlarged by the M290A mutation. The motion through which Phe-382 enters the N-pocket (to yield a structure like that shown in Fig. 1_B_) principally involves a change of its χ1 angle from approximately −60° to 60°. This structure is taken from simulation 4a after 2 ns. The conformation of Asp-381 and Phe-382 before Phe-382 enters the N-pocket is shown in a transparent rendering.
Fig. 3.
Conformational change involved in the DFG flip. All simulation structures shown are taken from the same simulation of the DFG flip (6b). The active Abl crystal structure (PDB entry 2F4J) is shown in pink. (A) A DFG-in to DFG-out flip. As indicated by the arrows, starting from conformation 1 (the active structure), Asp-381 leaves the ATP-binding site by the C-lobe (lower) side, whereas Phe-382 enters from above via the N-pocket. Also shown is the displacement of helix αC accompanying the DFG flip. Conformations 2, 3 and 4 are taken from the simulation at 1, 75, and 122 ns, respectively. In the interest of clarity, Asp-381 in conformation 3 is not shown (Asp-381 adopts a very similar position in conformations 3 and 4). (B) Active Abl (pink) compared with a conformation taken from the simulation (purple) showing a typical displacement of helix αC and conformational change of the DFG main chain before the DFG flip. The significant difference between the 2 helix-αC conformations reflects in part the twist and hinge motion of the N-lobe relative to the C-lobe.
Fig. 4.
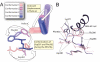
pH dependence of Abl-imatinib binding. (A) The on-rate constant of imatinib binding to wild-type Abl and to 3 mutants as a function of pH, as measured by stopped-flow tryptophan fluorescence assays. The error bars indicate the uncertainty of the linear regression fit to the observed rates from which the on-rate constants are obtained. (B) The relative population of DFG-out estimated according to Eq. 1. The similarity between the model results for _P_out and the experimental _k_on values supports our interpretation of the experimental data and the factors underlying DFG flips. (C) The observed fluorescence-decay rates for Abl-imatinib binding rates at 5 different pH values (virtually identical at pH 8.5 and pH 9). The solid lines are obtained from independent 2-parameter fits using the proposed kinetic scheme (see
SI Text
for details). (D) The dasatinib binding rate constants for wild-type Abl measured at 5 different pH values show only weak pH dependence. The lines are linear fits to the data.
References
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. - PubMed
- Cohen P. Protein kinases—The major drug targets of the twenty-first century? Nat Rev Drug Discov. 2002;1:309–315. - PubMed
- Zheng JH, et al. Crystal structure of the catalytic subunit of cAMP-dependent protein-kinase complexed with MgATP and peptide inhibitor. Biochem. 1993;32:2154–2161. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous