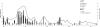Molecular epidemiology of norovirus gastroenteritis investigated using samples collected from children in Tunisia during a four-year period: detection of the norovirus variant GGII.4 Hunter as early as January 2003 - PubMed (original) (raw)
Molecular epidemiology of norovirus gastroenteritis investigated using samples collected from children in Tunisia during a four-year period: detection of the norovirus variant GGII.4 Hunter as early as January 2003
Khira Sdiri-Loulizi et al. J Clin Microbiol. 2009 Feb.
Abstract
Human noroviruses (NoVs) cause epidemic and endemic acute gastroenteritis in children and adults. To study the prevalence and genetic diversity of NoV in children in Tunisia, a total of 788 fecal samples were collected during a 4-year period in the region of Monastir, from children 12 years of age or younger, hospitalized or presenting in dispensaries with symptoms of acute gastroenteritis. NoV was detected by reverse transcription-PCR and confirmed by sequence analysis. This is the first report that describes the molecular epidemiology of NoV in Tunisian children: NoVs were characterized as the causative agent in 128 (16.2%) of the samples. Fourteen samples contained a mixture of two NoVs, and 33 samples were coinfected with additional enteric viruses. Eight distinct NoV genotypes were detected (GGI.2, GGI.4, GGII.1, GGII.4, GGII.8, GGII.14, GGIIb/GGII.2, and GGIIb/GGII.3). GGII.4 was the most prevalent genotype, accounting for 83 (64.8%) cases. Interestingly the GGII.4 variant Hunter, described as spreading all over the world in 2004, was found in Tunisia as early as January 2003. The delay of 1 year between the isolation in Tunisia and the worldwide emergence is somewhat surprising, considering the importance of the contacts between North Africa and Europe particularly. Nevertheless, this illustrates the idea that sporadic gastroenteritis cases may be a reservoir for emerging epidemic NoV strains.
Figures
FIG. 1.
Monthly distribution of NoV genotypes and numbers of samples tested in Tunisian children between January 2003 and April 2007. J through D, January through December, respectively.
FIG. 2.
Phylogenetic analysis based on the partial nucleotide sequences (285 bp) of the RNA-dependent RNA polymerase coding gene of the NoV strains associated with pediatric gastroenteritis in Monastir, Tunisia, between January 2003 and April 2007. The tree was constructed using UPGMA clustering. GenBank accession numbers for reference strains in this figure are as follows: Hunter virus, DQ078794; Southampton virus, L07418; Chiba virus, AB042808; Amsterdam virus, AB025907; GGIIb virus, AJ487794; Arg 320 virus, AF190817; Farmington Hills virus, AY502023. For more clarity, the sequences that present 100% nucleotide identity are not all represented on the tree. Only some of them, representative of each genotype, are indicated. In the same way, only some representative sequences were submitted to GenBank. Their GenBank accession numbers are indicated in Materials and Methods, and they are marked with asterisks in the tree. The percent bootstrap values in which the major groupings were observed among 100 replicates are indicated in each branch.
FIG. 3.
Phylogenetic analysis based on the partial nucleotide sequences (302 bp) of the capsid coding gene of NoV isolates detected in Tunisia. Reference strains of NoV were selected from the GenBank database under the following accession numbers: Farmington Hills virus, AY502023; Hunter virus, DQ078794; Amsterdam virus, AF195848; Hawaii virus, U07611; Melksham virus, X81879; Arg 320 virus, AF190817; M7/1999/US virus, AY130761. The GenBank accession numbers of strains of this study are indicated in the text, and they are marked with asterisks in the tree. The numbers in the branches indicate the bootstrap values.
FIG. 4.
Phylogenetic tree constructed from complete amino acid sequences of ORF2 of the seven Tunisian isolates and the GGII.4 representative strains from GenBank. For the samples in this study, accession numbers are as follows: EU916955 (Monastir-3968/2004), EU916956 (Monastir-8655/2007), EU916957 (Monastir-127/2003), EU916958 (Monastir-113/2003), EU916959 (Monastir-529/2003), EU916960 (Monastir-493/2003), and EU916961 (Monastir-715/2003). The numbers on each branch indicate the bootstrap values for the clusters supported by that branch.
Similar articles
- Detection of the norovirus variants GGII.4 hunter and GGIIb/hilversum in Italian children with gastroenteritis.
Ramirez S, De Grazia S, Giammanco GM, Milici M, Colomba C, Ruggeri FM, Martella V, Arista S. Ramirez S, et al. J Med Virol. 2006 Dec;78(12):1656-62. doi: 10.1002/jmv.20751. J Med Virol. 2006. PMID: 17063517 - Prevalence and genetic diversity of norovirus infection in Tunisian children (2007-2010).
Hassine-Zaafrane M, Sdiri-Loulizi K, Kaplon J, Salem IB, Pothier P, Aouni M, Ambert-Balay K. Hassine-Zaafrane M, et al. J Med Virol. 2013 Jun;85(6):1100-10. doi: 10.1002/jmv.23552. Epub 2013 Mar 26. J Med Virol. 2013. PMID: 23532785 - Prevalence and molecular epidemiology of noroviruses detected in outbreak and sporadic cases of acute gastroenteritis in Bulgaria.
Mladenova Z, Korsun N, Geonova T, Di Bartolo I, Fiore L, Ruggeri FM; Norovirus Study Group. Mladenova Z, et al. J Med Virol. 2008 Dec;80(12):2161-8. doi: 10.1002/jmv.21307. J Med Virol. 2008. PMID: 19040294 - Molecular epidemiology of noroviruses associated with acute sporadic gastroenteritis in children: global distribution of genogroups, genotypes and GII.4 variants.
Hoa Tran TN, Trainor E, Nakagomi T, Cunliffe NA, Nakagomi O. Hoa Tran TN, et al. J Clin Virol. 2013 Mar;56(3):185-93. doi: 10.1016/j.jcv.2012.11.011. Epub 2012 Dec 5. J Clin Virol. 2013. PMID: 23218993 Review. - A decade of norovirus disease risk among older adults in upper-middle and high income countries: a systematic review.
Lindsay L, Wolter J, De Coster I, Van Damme P, Verstraeten T. Lindsay L, et al. BMC Infect Dis. 2015 Oct 14;15:425. doi: 10.1186/s12879-015-1168-5. BMC Infect Dis. 2015. PMID: 26467099 Free PMC article. Review.
Cited by
- Genogroup IIb norovirus infections and association with enteric symptoms in a neonatal nursery in southern India.
Menon VK, George S, Ramani S, Illiayaraja J, Sarkar R, Jana AK, Kuruvilla KA, Kang G. Menon VK, et al. J Clin Microbiol. 2010 Sep;48(9):3212-5. doi: 10.1128/JCM.02510-09. Epub 2010 Jul 14. J Clin Microbiol. 2010. PMID: 20631107 Free PMC article. - Preadaptation of pandemic GII.4 noroviruses in unsampled virus reservoirs years before emergence.
Ruis C, Lindesmith LC, Mallory ML, Brewer-Jensen PD, Bryant JM, Costantini V, Monit C, Vinjé J, Baric RS, Goldstein RA, Breuer J. Ruis C, et al. Virus Evol. 2020 Nov 21;6(2):veaa067. doi: 10.1093/ve/veaa067. eCollection 2020 Jul. Virus Evol. 2020. PMID: 33381305 Free PMC article. - Rotavirus and norovirus infections among acute gastroenteritis children in Morocco.
El Qazoui M, Oumzil H, Baassi L, El Omari N, Sadki K, Amzazi S, Benhafid M, El Aouad R. El Qazoui M, et al. BMC Infect Dis. 2014 Jun 3;14:300. doi: 10.1186/1471-2334-14-300. BMC Infect Dis. 2014. PMID: 24894194 Free PMC article. - The epidemiology of Norovirus in the Middle East and North Africa (MENA) region: a systematic review.
Kreidieh K, Charide R, Dbaibo G, Melhem NM. Kreidieh K, et al. Virol J. 2017 Nov 10;14(1):220. doi: 10.1186/s12985-017-0877-3. Virol J. 2017. PMID: 29126448 Free PMC article. Review. - Etiology of Acute Diarrhea in Tunisian Children with Emphasis on Diarrheagenic Escherichia coli: Prevalence and Identification of E. coli Virulence Markers.
Ben Salem-Ben Nejma I, Hassine Zaafrane M, Hassine F, Sdiri-Loulizi K, Ben Said M, Aouni M, Mzoughi R. Ben Salem-Ben Nejma I, et al. Iran J Public Health. 2014 Jul;43(7):947-60. Iran J Public Health. 2014. PMID: 25909062 Free PMC article.
References
- Al-Mashhadani, M. N., O. Nakagomi, W. Dove, H. Ahmed, T. Nakagomi, C. A. Hart, and N. A. Cunliffe. 2008. Norovirus gastroenteritis among children in Iraqi Kurdistan. J. Med. Virol. 80506-509. - PubMed
- Armah, G. E., C. I. Gallimore, F. N. Binka, R. H. Asmah, J. Green, U. Ugoji, F. Anto, D. W. G. Brown, and J. J. Gray. 2006. Characterisation of norovirus strains in rural Ghanaian children with acute diarrhoea. J. Med. Virol. 781480-1485. - PubMed
- Bon, F., K. Ambert-Balay, H. Giraudon, J. Kaplon, S. Le Guyader, M. Pommepuy, A. Gallay, V. Vaillant, H. de Valk, R. Chikhi-Brachet, A. Flahaut, P. Pothier, and E. Kohli. 2005. Molecular epidemiology of caliciviruses detected in sporadic and outbreak cases of gastroenteritis in France from December 1998 to February 2004. J. Clin. Microbiol. 434659-4664. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical