DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45 - PubMed (original) (raw)
DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45
Kunal Rai et al. Cell. 2008.
Abstract
Evidence for active DNA demethylation in vertebrates is accumulating, but the mechanisms and enzymes remain unclear. Using zebrafish embryos we provide evidence for 5-methylcytosine (5-meC) removal in vivo via the coupling of a 5-meC deaminase (AID, which converts 5-meC to thymine) and a G:T mismatch-specific thymine glycosylase (Mbd4). The injection of methylated DNA into embryos induced a potent DNA demethylation activity, which was attenuated by depletion of AID or the non enzymatic factor Gadd45. Remarkably, overexpression of the deaminase/glycosylase pair AID/Mbd4 in vivo caused demethylation of the bulk genome and injected methylated DNA fragments, likely involving a G:T intermediate. Furthermore, AID or Mbd4 knockdown caused the remethylation of a set of common genes. Finally, Gadd45 promoted demethylation and enhanced functional interactions between deaminase/glycosylase pairs. Our results provide evidence for a coupled mechanism of 5-meC demethylation, whereby AID deaminates 5-meC, followed by thymine base excision by Mbd4, promoted by Gadd45.
Figures
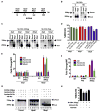
Figure 1. Involvement of Cytidine Deaminases in DNA Demethylation in Zebrafish
(A) Schematic of the M-DNA (Methylated DNA) fragment injected into fertilized embryos at the single-cell stage. The four HpaII/MspI sites are indicated. HpaII-resistant (methylated CmeCGG) and HpaII-cleaved species (unmethylated) run at ~750 bp and ~250 bp, respectively (B). (B) M-DNA methylation status was assessed by HpaII susceptibility, with cutting observed on unmethylated (U) (lane 2) but not methylated (Me) DNA (lane 2). MspI-digested (lanes 3 and 6) and uncut DNA (lanes 1 and 4) served as controls. Fragments were detected by Southern blot analysis probed with full-length M-DNA probe. (C) M-DNA methylation status during development. Total DNA was isolated at the time points shown from embryos injected at the single-cell stage with M-DNA (5 pg or 200 pg), and treated as in (B). M-DNA induced demethylation peaks at ~13 hpf (Compare lanes 2, 5, 8, and 11). (D) LC-MS quantitation of 5-meC content of the bulk genome (normalized to total deoxyguanosine content) in genomic DNA isolated from embryos (hpf indicated) injected with methylated M-DNA or unmethylated U-DNA at the single-cell stage. (E and F) qRT-PCR determinations from embryos injected with M-DNA (E), and at different fragment concentrations (F). (G and H) Methylation status of M-DNA assessed by HpaII digestion and Southern blotting (G), or LC-MS quantitation of total 5-MeC (H) in total genomic DNA isolated from embryos at 13 hpf, injected at the single-cell stage with M-DNA (200 pg) and morpholinos as indicated. Lanes 1, 7, and 13 correspond to wild-type sample. AAAmm refers to a set of three control morpholinos against AID (4 pg), Apobec2a (4 pg), and Apobec2b (2 pg) (AAA), which each contain five mismatched (mm) bases (of 25 total to prevent binding) relative to the efficacious morpholino (same amount as controls). For HpaII/MspI susceptibility, one representative of at least three biological repeats is shown. LC-MS measurements; two biological replicates. Asterisks (*) depict statistical significance (p < 0.05). Error bars: +/− one standard deviation.
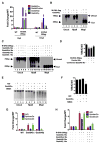
Figure 2. Overexpression of a Deaminase/Glycosylase Pair Elicits DNA Demethylation
(A–C) Methylation status assessed by HpaII digestion of total genomic DNA (A), LC-MS quantitation ([B] upper panel), HpaII digestion of M-DNA (Southern analysis) ([B] lower panel), and bisulphite sequencing of M-DNA (C). Lanes 1, 7, and 13 in (A) and lane 1 in (B) correspond to wild-type sample. For (B), M-DNA was injected at 5 pg, below the threshold level for eliciting demethylation on its own (See Figures 1C and 1D). For (C), twenty clones were subjected to bisulphite sequencing, and the methylation status of each HpaII/MspI (CCGG) site reported as a percentage of total sites tested. (D) Repeat elements from DNA isolated from embryos (13 hpf) injected at the single-cell stage with RNA encoding wild-type AID, along with MBD4 wild-type mRNA. For each experiment, one representative of at least three biological repeats is shown except in LC-MS measurement where graph is prepared from values of two biological replicates. Asterisks (*) depict statistical significance (p < 0.05). Error bars are ± one standard deviation.
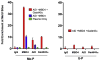
Figure 3. A PCR Strategy to Detect a G:T Intermediate
(A) Schematic of the PCR reaction for thymine (CmeCGG > CTGG) detection at M-DNA HpaII/MspI sites using an A-tailed primer (only 3 of the ~22 bases shown) with an adenosine at the 3′ end. (B) Detection of a G:T mismatch on M-DNA by PCR. M-DNA, AID mRNA, and RNA encoding either wild-type or catalytically inactive hMbd4 (D560A) was injected at the single-cell stage and assessed at 13 hpf.
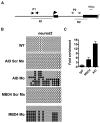
Figure 4. Gadd45 Proteins Promote Demethylation and Selectively Upregulate Deaminases
(A) Gadd45 family members are upregulated by M-DNA, assessed by RT-PCR. (B) Gadd45_α_ induces moderate demethylation of M-DNA as detected by HpaII digestion and subsequent Southern blotting. M-DNA is injected at 5 pg which does not induce demethylation on its own. Lanes 1, 4, and 7 correspond to wild-type sample. (C and D) Lowering the levels of Gadd45 family members via morpholino injection attenuates de-methylation. Demethylation of 5-methylcytosine as assessed by HpaII digestion and Southern blotting (C), or LC-MS quantitation of 5-meC (D) in total genomic DNA isolated from 13 hpf old embryos injected at the single-cell stage with M-DNA alone (200 pgs) or along with morpholinos as shown. Lanes 1, 7 and 13 correspond to wild-type sample. Combined Gadd45 Mo refers to the combination of morpholinos to all four Gadd45 family members tested (α,α_-like, β, and γ; 2 pg each). (E and F) Synergy among AID, hMbd4, and Gadd45 for demethylation. Methylation status of total genomic DNA from embryos (13 hpf) was assessed by HpaII susceptibility (E) or by LC-MS quantitation of global 5-methylcytosine levels (F) injected at single-cell stage with mRNAs encoding factors as indicated. Lanes 1, 5, and 9 correspond to wild-type sample. Note: AID, MBD4, and Gadd45_α were injected at subthreshold amounts (at 25 pgs each), levels which are not sufficient to induce demethylation alone. (G and H) Quantitative RT-PCR for deaminase family members (assessed at 13 hpf) injected at single-cell stage with mRNA encoding Gadd45_α_ or Gadd45_β_ (F) and M-DNA (200 pg) alone or with morpholinos as shown (G). For each experiment, one representative of at least three biological repeats is shown except in LC-MS measurement where graph is prepared from values of two biological replicates. Asterisks (*) depict statistical significance (p < 0.05). Error bars: are ± one standard deviation.
Figure 5. Gadd45_α_ Promotes Recruitment of AID and MBD4 to Methylated Regions of a Plasmid In Vivo
Enrichment of AID, MBD4, and Gadd45_α_ on pCMV-Luc, which contains both methylated (Me) and unmethylated (U) regions. ChIP experiments with extracts from embryos (12 hpf) injected at the single-cell stage with V5-tagged AID, HA-tagged hMbd4, His-tagged Gadd45_α_ and in vitro-methylated (by HpaII methylase) pCMV-Luc (Me-P). Y–axis values represent the ratio of enrichment on a DNA segment containing in vitro methylated CmeCGG sites to enrichment on a site (also on pCMV-Luc) containing no CCGG elements. Me-P and U-P on axis depict methylated and unmethylated plasmid, respectively. Graph shows one representative experiment of three biological repeats. Error bars: are ± one standard deviation.
Figure 6. AID and hMbd4 Occupy the neurod2 Promoter and Affect the Methylation Status at 80% Epiboly
(A) Schematic of the neurod2 promoter and start site region. R1 and R2 show regions of bisulfite sequencing (Results shown for only R1; R2 remains unmethylated and unaffected). P1 and P2 depict the amplicons used for ChIP determinations. (B) Bisulphite sequencing of the R1 region of the neurod2 promoter in the WT (uninjected) animals or animals injected with morpholinos (all 2 pg) as shown. Scr Mo; a control morpholino where the base composition is maintained, but the order scrambled. (C) Enrichment of AID and hMbd4 at neurod2 (P1 versus P2). ChIP experiments with extracts from embryos at 80% epiboly, which were initially injected at the single-cell stage with V5-tagged AID and HA-tagged hMbd4. Graph shows one representative biological experiment (two biological repeats), with the average of three technical replicates shown. Error bars are ± one standard deviation.
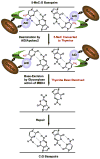
Figure 7. Model for 5-meC Demethylation
Demethylation may occur through a two-step coupled enzymatic process, promoted by Gadd45. The first enzymatic step involves deamination of 5-meC by AID (amine group removed, in blue), generating a thymine product and a G:T mismatch. The second step involves thymine base removal by Mdb4, generating an abasic site. As the transient G:T intermediate is not detected in cells with active Mbd4, but is with catalytically inactive Mbd4, the thymine is likely rapidly removed, suggesting a coupling between deaminase and glycosylase activity. Gadd45 may promote functional or physical interactions between AID and Mbd4 at the site of demethylation. Mbd4 may couple with a lyase to help promote base replacement through base excision repair (neither shown nor addressed). Targeting of AID/Mbd4 may be promoted by recognition of the 5-meCpG (methyl group in red), or through other mechanisms (data not shown).
Comment in
- DNA Cytosine demethylation: are we getting close?
Jiricny J, Menigatti M. Jiricny J, et al. Cell. 2008 Dec 26;135(7):1167-9. doi: 10.1016/j.cell.2008.12.008. Cell. 2008. PMID: 19109887
Similar articles
- No evidence for AID/MBD4-coupled DNA demethylation in zebrafish embryos.
Shimoda N, Hirose K, Kaneto R, Izawa T, Yokoi H, Hashimoto N, Kikuchi Y. Shimoda N, et al. PLoS One. 2014 Dec 23;9(12):e114816. doi: 10.1371/journal.pone.0114816. eCollection 2014. PLoS One. 2014. PMID: 25536520 Free PMC article. - DNA Cytosine demethylation: are we getting close?
Jiricny J, Menigatti M. Jiricny J, et al. Cell. 2008 Dec 26;135(7):1167-9. doi: 10.1016/j.cell.2008.12.008. Cell. 2008. PMID: 19109887 - 5-Methylcytosine DNA glycosylase activity is also present in the human MBD4 (G/T mismatch glycosylase) and in a related avian sequence.
Zhu B, Zheng Y, Angliker H, Schwarz S, Thiry S, Siegmann M, Jost JP. Zhu B, et al. Nucleic Acids Res. 2000 Nov 1;28(21):4157-65. doi: 10.1093/nar/28.21.4157. Nucleic Acids Res. 2000. PMID: 11058112 Free PMC article. - Active DNA demethylation mediated by DNA glycosylases.
Zhu JK. Zhu JK. Annu Rev Genet. 2009;43:143-66. doi: 10.1146/annurev-genet-102108-134205. Annu Rev Genet. 2009. PMID: 19659441 Free PMC article. Review. - DNA demethylation pathways: Additional players and regulators.
Bochtler M, Kolano A, Xu GL. Bochtler M, et al. Bioessays. 2017 Jan;39(1):1-13. doi: 10.1002/bies.201600178. Epub 2016 Nov 16. Bioessays. 2017. PMID: 27859411 Review.
Cited by
- Decrease in cytosine methylation at CpG island shores and increase in DNA fragmentation during zebrafish aging.
Shimoda N, Izawa T, Yoshizawa A, Yokoi H, Kikuchi Y, Hashimoto N. Shimoda N, et al. Age (Dordr). 2014 Feb;36(1):103-15. doi: 10.1007/s11357-013-9548-5. Epub 2013 Jun 5. Age (Dordr). 2014. PMID: 23736955 Free PMC article. - Global DNA hypomethylation in peripheral blood leukocytes as a biomarker for cancer risk: a meta-analysis.
Woo HD, Kim J. Woo HD, et al. PLoS One. 2012;7(4):e34615. doi: 10.1371/journal.pone.0034615. Epub 2012 Apr 11. PLoS One. 2012. PMID: 22509334 Free PMC article. - Zebrafish (Danio rerio) life-cycle exposure to chronic low doses of ethinylestradiol modulates p53 gene transcription within the gonads, but not NER pathways.
Soares J, Castro LF, Reis-Henriques MA, Monteiro NM, Santos MM. Soares J, et al. Ecotoxicology. 2012 Jul;21(5):1513-22. doi: 10.1007/s10646-012-0905-4. Epub 2012 Apr 28. Ecotoxicology. 2012. PMID: 22543959 - Chromatin Memory in the Development of Human Cancers.
Yao Y, Des Marais TL, Costa M. Yao Y, et al. Gene Technol. 2014 Aug 11;3(2):114. Gene Technol. 2014. PMID: 25606572 Free PMC article. - The expression of mouse double minute 2 homolog and P73 had no correlation with growth arrest DNA damage-inducible gene 45α in patients with non-small-cell lung carcinoma: A STROBE-compliant study.
Wang B, Liang C, Liu H, Lin J, Wang B, Fan K, Ren Z, Wang B, Li T, Qi K, Tian X. Wang B, et al. Medicine (Baltimore). 2019 Dec;98(51):e17944. doi: 10.1097/MD.0000000000017944. Medicine (Baltimore). 2019. PMID: 31860949 Free PMC article.
References
- Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, Niehrs C. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. - PubMed
- Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. - PubMed
- Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol. 2003;4:235–240. - PubMed
- Conticello SG, Langlois MA, Yang Z, Neuberger MS. DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv Immunol. 2007;94:37–73. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA116674/CA/NCI NIH HHS/United States
- P30 CA016056/CA/NCI NIH HHS/United States
- CA16056/CA/NCI NIH HHS/United States
- R01 HD058506-01/HD/NICHD NIH HHS/United States
- R01 CA116674-03/CA/NCI NIH HHS/United States
- R01 HD058506/HD/NICHD NIH HHS/United States
- CA24014/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases