Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II - PubMed (original) (raw)
Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II
Thomas S Ream et al. Mol Cell. 2009.
Abstract
In addition to RNA polymerases I, II, and III, the essential RNA polymerases present in all eukaryotes, plants have two additional nuclear RNA polymerases, abbreviated as Pol IV and Pol V, that play nonredundant roles in siRNA-directed DNA methylation and gene silencing. We show that Arabidopsis Pol IV and Pol V are composed of subunits that are paralogous or identical to the 12 subunits of Pol II. Four subunits of Pol IV are distinct from their Pol II paralogs, six subunits of Pol V are distinct from their Pol II paralogs, and four subunits differ between Pol IV and Pol V. Importantly, the subunit differences occur in key positions relative to the template entry and RNA exit paths. Our findings support the hypothesis that Pol IV and Pol V are Pol II-like enzymes that evolved specialized roles in the production of noncoding transcripts for RNA silencing and genome defense.
Figures
Figure 1. Relationships of Arabidopsis Pol II, IV, and V Subunits to E. coli, Archaeai, and Yeast RNA Pol II Subunits
Numbers indicate percent protein coverage represented by peptides unique to that protein. “*” indicates that all peptides match both closely related proteins. Unshaded numbers represent alternate subunits detected at trace levels relative to the predominant subunit.
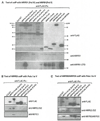
Figure 2. Verification of Pol V Subunit Associations
(A) Pol V includes subunits shared with other polymerases as well as a unique RPB5 family variant. Pol I, II, III, IV, and V were immunoprecipitated by virtue of FLAG-tagged catalytic subunits alongside NRPE6a, NRPE8b, NRPE10, NRPE11, and NRPE5 FLAG-tagged subunits. Duplicate immunoblots were probed with anti-FLAG, anti-NRPE1, anti-NRPE2/NRPD2 (abbreviated anti-NRPE2/D2), or an antibody recognizing the second-largest subunits of Pol I, II, or III. The two panels in the top row are from the same blot but focus on different size ranges. (B) Control immunoblot showing that the multiple high-molecular-mass bands characteristic of NRPE1 are lost in an nrpe1 null mutant (allele nrpd1b-11), indicating that the antibody is specific for NRPE1. (C) NRPE6b and NRPE9a are subunits of Pol V as well as Pol I, II, or III. Immunoprecipitation and immunoblot detection was as in (A). NRPE5 and NRPB2 immunoprecipitations serve as controls for Pol V and Pol II, respectively. “*” denotes a nonspecific band detected by the anti-FLAG antibody. (D) Phylogenetic tree based on a CLUSTALW alignment of Arabidopsis RPB9-like proteins with the RPB9 (Pol II), RPC11 (Pol III), and RPA12 (Pol I) subunit equivalents of yeast. (E) NRPE8a and NRPE6a associate with Pol V. Immunoprecipitation and immunoblot detection was as in (A). NRPE5 and NRPB7 serve as controls for Pol V and Pol II, respectively.
Figure 3. Pol V Utilizes a Distinct RPB3 Variant, NRPE3b, as well as an NRPE3a Variant Corresponding to the Pol II NRPB3 Subunit
(A) Alignment of the two Arabidopsis RPB3 family proteins with yeast RPB3. (B) HA-tagged NRPE3a/NRPB3 and NRPE3b were Immunoprecipitated and resulting immunoblots were probed using the indicated antibodies.
Figure 4. ColP Tests of Pol V, IV, and II Subunit Associations
(A) Pol I, II, III, IV, and V were immunoprecipitated by virtue of FLAG-tagged catalytic subunits along-side immunoprecipitated NRPE6a, NRPE8b, NRPE9a, NRPE10, NRPE11, and NRPE3b FLAG-tagged subunits. Duplicate immunoblots were probed with anti-FLAG, anti-NRPD1 (Pol IV), or anti-NRPB1-CTD (Pol II). The two panels in the top row show different exposures of the same blot, focused on different size ranges. (B) Pol I, II, III, IV, and V were immunoprecipitated using the indicated FLAG-tagged subunits and probed with anti-FLAG, anti-NRPE5, or anti-NRPE2/NRPD2. (C) Immunoprecipitation and immunoblotting using the indicated antibodies were as in (B).
Figure 5. nrpe5 Mutants Are Defective in RNA-Directed DNA Methylation and Retrotransposon Silencing
(A) Phylogenetic tree based on a CLUSTALW alignment of the five full-length RPB5-like proteins in Arabidopsis with the RPB5 subunits of yeast and human. (B) Locations of T-DNA insertions in the nrpb5-1/nrpe5-1 and nrpe5-1 alleles. Black boxes represent exons, black bars represent introns, and gray bars represent 5′ and 3′UTRs. (C) nrpe5-1 homozygous mutant plants display a delay in flowering under short-day conditions (8 hr light, 16 hr dark). The mean (±SEM) number of rosette leaves when the floral bolt reached 10 cm is graphed. All mutants are significantly different from wild-type based on a Student’s t test (p < 0.05). (D) Methylation-sensitive Southern blot analyses of wild-type, nrpe1, nrpd2/nrpd2, and nrpe5 mutants and three different nrpe5, 35S:FLAG-NRPE5 transgenic lines. Genomic DNA was digested with either _Hpa_II (left, reports on meCG) or _Hae_III (right, reports on meCNN) and probed for 5S rDNA repeats. Images for the _Hpa_II or _Hae_III digests are from the same exposures of the same Southern blots; the black vertical lines separate groups of lanes whose order was rearranged for clarity of presentation. (E and F) PCR-based methylation assay of AtSN1 and AtSN2 family retroelements. Genomic DNA was digested with _Hae_III and subjected to PCR using AtSN1, AtSN2-1, or control primers that amplify sequences lacking _Hae_III sites (At2g19920 in the case of [B], and an AtSN2 family element lacking _Hae_III sites in the case of [C]). Diagrams show the relative positions of the primers flanking the _Hae_III sites. (G) RT-PCR detection of AtSN1 and actin transcripts. (H) Small RNA blot analysis. Blots were probed for siRNAs corresponding to 45S or 5S rRNA genes, Copia or AtSN1 transposons, and miRNA 173 or _trans_-acting siRNA 255.
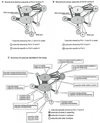
Figure 6. Comparison of RNA Polymerase Subunits in Pol II, IV, and V
(A) Subunits that are unique to Pol IV and/or Pol V compared to Pol II are shown in blue. Subunits common to Pol II, IV, and V are shown in green. The subunit interaction model is based on the yeast Pol II crystal structure (Armache et al., 2005; Cramer et al., 2001; Sampath et al., 2008). The thickness of lines connecting the subunits is proportional to the number of contacts. (B) Subunits that are unique to Pol V are shown in blue. Subunits common to Pol IV and Pol V are shown in green. The half-blue, half-green shading of the third-largest subunit reflects the fact that Pol V uses the NRPE3b variant that is not used appreciably by Pol IV in addition to the NRPE3a/NRPD3 variant that predominates in Pol IV. (C) Summary of the Arabidopsis genes that encode Pol II, IV, or V subunits.
Similar articles
- Metal A and metal B sites of nuclear RNA polymerases Pol IV and Pol V are required for siRNA-dependent DNA methylation and gene silencing.
Haag JR, Pontes O, Pikaard CS. Haag JR, et al. PLoS One. 2009;4(1):e4110. doi: 10.1371/journal.pone.0004110. Epub 2009 Jan 1. PLoS One. 2009. PMID: 19119310 Free PMC article. - NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation.
He XJ, Hsu YF, Pontes O, Zhu J, Lu J, Bressan RA, Pikaard C, Wang CS, Zhu JK. He XJ, et al. Genes Dev. 2009 Feb 1;23(3):318-30. doi: 10.1101/gad.1765209. Genes Dev. 2009. PMID: 19204117 Free PMC article. - Functional consequences of subunit diversity in RNA polymerases II and V.
Tan EH, Blevins T, Ream TS, Pikaard CS. Tan EH, et al. Cell Rep. 2012 Mar 29;1(3):208-14. doi: 10.1016/j.celrep.2012.01.004. Cell Rep. 2012. PMID: 22550619 Free PMC article. - Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing.
Haag JR, Pikaard CS. Haag JR, et al. Nat Rev Mol Cell Biol. 2011 Jul 22;12(8):483-92. doi: 10.1038/nrm3152. Nat Rev Mol Cell Biol. 2011. PMID: 21779025 Review. - Molecular mechanisms of the RNA polymerases in plant RNA-directed DNA methylation.
Xie G, Du X, Hu H, Du J. Xie G, et al. Trends Biochem Sci. 2024 Mar;49(3):247-256. doi: 10.1016/j.tibs.2023.11.005. Epub 2023 Dec 9. Trends Biochem Sci. 2024. PMID: 38072749 Review.
Cited by
- The RNA silencing enzyme RNA polymerase v is required for plant immunity.
López A, Ramírez V, García-Andrade J, Flors V, Vera P. López A, et al. PLoS Genet. 2011 Dec;7(12):e1002434. doi: 10.1371/journal.pgen.1002434. Epub 2011 Dec 29. PLoS Genet. 2011. PMID: 22242006 Free PMC article. - An RdDM-independent function of Pol V transcripts in gene regulation and plant defence.
Yuan Y, Liu Y, Han L, Li Y, Qi Y. Yuan Y, et al. Nat Plants. 2024 Oct;10(10):1562-1575. doi: 10.1038/s41477-024-01774-0. Epub 2024 Aug 26. Nat Plants. 2024. PMID: 39187700 - Domains rearranged methyltransferase3 controls DNA methylation and regulates RNA polymerase V transcript abundance in Arabidopsis.
Zhong X, Hale CJ, Nguyen M, Ausin I, Groth M, Hetzel J, Vashisht AA, Henderson IR, Wohlschlegel JA, Jacobsen SE. Zhong X, et al. Proc Natl Acad Sci U S A. 2015 Jan 20;112(3):911-6. doi: 10.1073/pnas.1423603112. Epub 2015 Jan 5. Proc Natl Acad Sci U S A. 2015. PMID: 25561521 Free PMC article. - A conserved transcriptional regulator is required for RNA-directed DNA methylation and plant development.
He XJ, Hsu YF, Zhu S, Liu HL, Pontes O, Zhu J, Cui X, Wang CS, Zhu JK. He XJ, et al. Genes Dev. 2009 Dec 1;23(23):2717-22. doi: 10.1101/gad.1851809. Epub 2009 Nov 10. Genes Dev. 2009. PMID: 19903758 Free PMC article. - Epigenetic regulation in plants.
Pikaard CS, Mittelsten Scheid O. Pikaard CS, et al. Cold Spring Harb Perspect Biol. 2014 Dec 1;6(12):a019315. doi: 10.1101/cshperspect.a019315. Cold Spring Harb Perspect Biol. 2014. PMID: 25452385 Free PMC article. Review.
References
- Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. - PubMed
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. - PubMed
- Armache KJ, Mitterweger S, Meinhart A, Cramer P. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J. Biol. Chem. 2005;280:7131–7134. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases