A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters - PubMed (original) (raw)
. 2008 Dec 26;32(6):878-87.
doi: 10.1016/j.molcel.2008.11.020.
Esther T Chan, Harm van Bakel, Lourdes Pena-Castillo, Desiree Tillo, Kyle Tsui, Clayton D Carlson, Andrea J Gossett, Michael J Hasinoff, Christopher L Warren, Marinella Gebbia, Shaheynoor Talukder, Ally Yang, Sanie Mnaimneh, Dimitri Terterov, David Coburn, Ai Li Yeo, Zhen Xuan Yeo, Neil D Clarke, Jason D Lieb, Aseem Z Ansari, Corey Nislow, Timothy R Hughes
Affiliations
- PMID: 19111667
- PMCID: PMC2743730
- DOI: 10.1016/j.molcel.2008.11.020
A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters
Gwenael Badis et al. Mol Cell. 2008.
Abstract
The sequence specificity of DNA-binding proteins is the primary mechanism by which the cell recognizes genomic features. Here, we describe systematic determination of yeast transcription factor DNA-binding specificities. We obtained binding specificities for 112 DNA-binding proteins representing 19 distinct structural classes. One-third of the binding specificities have not been previously reported. Several binding sequences have striking genomic distributions relative to transcription start sites, supporting their biological relevance and suggesting a role in promoter architecture. Among these are Rsc3 binding sequences, containing the core CGCG, which are found preferentially approximately 100 bp upstream of transcription start sites. Mutation of RSC3 results in a dramatic increase in nucleosome occupancy in hundreds of proximal promoters containing a Rsc3 binding element, but has little impact on promoters lacking Rsc3 binding sequences, indicating that Rsc3 plays a broad role in targeting nucleosome exclusion at yeast promoters.
Figures
Fig 1
Motifs identified in our study.
Fig 2. Comparison of motif representation and reproducibility of 8-mer profiles across platforms
(A) PWM scores (Granek and Clarke, 2005) for all possible 8-mers for the single motif with highest Pearson correlation to the PBM 8-mers, plotted against the Z-scores from the PBM. (B) CSI Z-scores (combined from up to four array spots containing the 8-mer) vs. Z-scores from PBM. Data are plotted as asinh values, which are similar to natural log, but return real values for negative numbers (by definition, half of all Z-scores are negative).
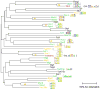
Fig 3. Similarity among C2H2 zinc finger motifs reflects DNA-binding domain sequence similarity
The phylogram tree was created using online EBI ClustalW with default settings. Our motifs are shown next to the gene names; inconsistent motifs from (MacIsaac et al., 2006) are shown for Stp4 and Yml081w. Yellow asterisks represent pairs arising from the WGD. Colors of protein names reflect our classifications of consistency with prior data: Green; known motif obtained; Red; discrepancy between our motif and that previously reported; Yellow, new motif but consistent with expectation based on homology; Blue, new unexpected motif.
Fig 4. Bias in the position of TF binding sequences in 5,015 promoters with well-defined TSS (Lee et al., 2007)
Motif scores (Granek and Clarke, 2005) were calculated for 8-bp windows, and high-scoring 8-mers were tallied along equivalent positions of all of the yeast promoter sequences using a cutoff selected to capture only the linear range of Z vs. PWM score in PBM experiments (cutoff values are given in Supplementary data). Background was calculated from the first 100 bases of yeast ORFs. TFs are sorted by relative enrichment between −125 and −75.
Fig 5. Rsc3 influences nucleosome occupancy at proximal promoters containing Rsc3 binding sites
(A) A segment of Chromosome XIII with a Rsc3 binding sequence (grey vertical line) that is depleted in wildtype but occupied in the rsc3-1 mutant. (B, C) Changes in promoter nucleosome occupancy profiles between rsc3-1 and a wildtype control for promoters containing Rsc3 binding sequences (C), or containing Reb1 binding sequences but not Rsc3 binding sequences (D). Promoters are sorted by the position of the highest scoring Rsc3 or Reb1 binding sequence location in the promoter, which is shown at left in panels B and C. Additional sites of equivalent PWM score are also indicated.
Fig 6. Comparison of the effects of mutations in essential DNA-binding proteins on nucleosome profiles at all promoters
Promoters are sorted by change in occupancy in the NFR. Locations of binding sequences for the mutated factor are illustrated at left, in tiling intervals matching those of the array, and shown as heat-maps. Relative transcript levels are illustrated at right. The rsc3-1 panel (upper left) also shows the change in relative enrichment in Rsc8-TAP ChIP-chip between the rsc3-1 and wildtype strains.
Similar articles
- Transcription factor binding site positioning in yeast: proximal promoter motifs characterize TATA-less promoters.
Erb I, van Nimwegen E. Erb I, et al. PLoS One. 2011;6(9):e24279. doi: 10.1371/journal.pone.0024279. Epub 2011 Sep 9. PLoS One. 2011. PMID: 21931670 Free PMC article. - Different roles for abf1p and a T-rich promoter element in nucleosome organization of the yeast RPS28A gene.
Lascaris RF, Groot E, Hoen PB, Mager WH, Planta RJ. Lascaris RF, et al. Nucleic Acids Res. 2000 Mar 15;28(6):1390-6. doi: 10.1093/nar/28.6.1390. Nucleic Acids Res. 2000. PMID: 10684934 Free PMC article. - Heat shock factor can activate transcription while bound to nucleosomal DNA in Saccharomyces cerevisiae.
Pederson DS, Fidrych T. Pederson DS, et al. Mol Cell Biol. 1994 Jan;14(1):189-99. doi: 10.1128/mcb.14.1.189-199.1994. Mol Cell Biol. 1994. PMID: 8264586 Free PMC article. - The pattern and evolution of yeast promoter bendability.
Tirosh I, Berman J, Barkai N. Tirosh I, et al. Trends Genet. 2007 Jul;23(7):318-21. doi: 10.1016/j.tig.2007.03.015. Epub 2007 Apr 6. Trends Genet. 2007. PMID: 17418911 Review. - Establishing nucleosome architecture and stability at promoters: Roles of pioneer transcription factors and the RSC chromatin remodeler.
Kubik S, Bruzzone MJ, Shore D. Kubik S, et al. Bioessays. 2017 May;39(5). doi: 10.1002/bies.201600237. Epub 2017 Mar 27. Bioessays. 2017. PMID: 28345796 Review.
Cited by
- Constitutive turnover of histone H2A.Z at yeast promoters requires the preinitiation complex.
Tramantano M, Sun L, Au C, Labuz D, Liu Z, Chou M, Shen C, Luk E. Tramantano M, et al. Elife. 2016 Jul 20;5:e14243. doi: 10.7554/eLife.14243. Elife. 2016. PMID: 27438412 Free PMC article. - Dry and wet approaches for genome-wide functional annotation of conventional and unconventional transcriptional activators.
Levati E, Sartini S, Ottonello S, Montanini B. Levati E, et al. Comput Struct Biotechnol J. 2016 Jun 29;14:262-70. doi: 10.1016/j.csbj.2016.06.004. eCollection 2016. Comput Struct Biotechnol J. 2016. PMID: 27453771 Free PMC article. Review. - Epigenetic pioneering by SWI/SNF family remodelers.
Ahmad K, Brahma S, Henikoff S. Ahmad K, et al. Mol Cell. 2024 Jan 18;84(2):194-201. doi: 10.1016/j.molcel.2023.10.045. Epub 2023 Nov 27. Mol Cell. 2024. PMID: 38016477 Review. - Timing of transcriptional quiescence during gametogenesis is controlled by global histone H3K4 demethylation.
Xu M, Soloveychik M, Ranger M, Schertzberg M, Shah Z, Raisner R, Venkatasubrahmanyan S, Tsui K, Gebbia M, Hughes T, van Bakel H, Nislow C, Madhani HD, Meneghini MD. Xu M, et al. Dev Cell. 2012 Nov 13;23(5):1059-71. doi: 10.1016/j.devcel.2012.10.005. Epub 2012 Nov 1. Dev Cell. 2012. PMID: 23123093 Free PMC article. - The yeast Snt2 protein coordinates the transcriptional response to hydrogen peroxide-mediated oxidative stress.
Baker LA, Ueberheide BM, Dewell S, Chait BT, Zheng D, Allis CD. Baker LA, et al. Mol Cell Biol. 2013 Oct;33(19):3735-48. doi: 10.1128/MCB.00025-13. Epub 2013 Jul 22. Mol Cell Biol. 2013. PMID: 23878396 Free PMC article.
References
- Angus-Hill ML, Schlichter A, Roberts D, Erdjument-Bromage H, Tempst P, Cairns BR. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol Cell. 2001;7:741–751. - PubMed
- Barrera LO, Ren B. The transcriptional regulatory code of eukaryotic cells--insights from genome-wide analysis of chromatin organization and transcription factor binding. Curr Opin Cell Biol. 2006;18:291–298. - PubMed
- Beer MA, Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–198. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01-GM072518/GM/NIGMS NIH HHS/United States
- T15LM007359/LM/NLM NIH HHS/United States
- R01 GM072518-02/GM/NIGMS NIH HHS/United States
- T15 LM007359/LM/NLM NIH HHS/United States
- R01 GM072518-03/GM/NIGMS NIH HHS/United States
- R01 GM069420-04/GM/NIGMS NIH HHS/United States
- R01 GM069420/GM/NIGMS NIH HHS/United States
- R01 GM072518/GM/NIGMS NIH HHS/United States
- R01 GM072518-01A1/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases