Genome-wide co-expression analysis in multiple tissues - PubMed (original) (raw)
Genome-wide co-expression analysis in multiple tissues
Ian C Grieve et al. PLoS One. 2008.
Abstract
Expression quantitative trait loci (eQTLs) represent genetic control points of gene expression, and can be categorized as cis- and trans-acting, reflecting local and distant regulation of gene expression respectively. Although there is evidence of co-regulation within clusters of trans-eQTLs, the extent of co-expression patterns and their relationship with the genotypes at eQTLs are not fully understood. We have mapped thousands of cis- and trans-eQTLs in four tissues (fat, kidney, adrenal and left ventricle) in a large panel of rat recombinant inbred (RI) strains. Here we investigate the genome-wide correlation structure in expression levels of eQTL transcripts and underlying genotypes to elucidate the nature of co-regulation within cis- and trans-eQTL datasets. Across the four tissues, we consistently found statistically significant correlations of cis-regulated gene expression to be rare (<0.9% of all pairs tested). Most (>80%) of the observed significant correlations of cis-regulated gene expression are explained by correlation of the underlying genotypes. In comparison, co-expression of trans-regulated gene expression is more common, with significant correlation ranging from 2.9%-14.9% of all pairs of trans-eQTL transcripts. We observed a total of 81 trans-eQTL clusters (hot-spots), defined as consisting of > or =10 eQTLs linked to a common region, with very high levels of correlation between trans-regulated transcripts (77.2-90.2%). Moreover, functional analysis of large trans-eQTL clusters (> or =30 eQTLs) revealed significant functional enrichment among genes comprising 80% of the large clusters. The results of this genome-wide co-expression study show the effects of the eQTL genotypes on the observed patterns of correlation, and suggest that functional relatedness between genes underlying trans-eQTLs is reflected in the degree of co-expression observed in trans-eQTL clusters. Our results demonstrate the power of an integrative, systematic approach to the analysis of a large gene expression dataset to uncover underlying structure, and inform future eQTL studies.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
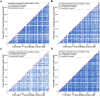
Figure 1. Scatter plots showing pairs of _cis_-eQTL genes with significantly correlated expression profiles by genetic map location.
Significantly correlated pairs of _cis_-eQTL genes identified in a) fat, b) kidney, c) adrenal and d) left ventricle are plotted according to genetic map location (cM), indicating significance or otherwise of correlation of strain distribution patterns (SDPs) at the peak of linkage. Colour coding indicates significance of correlation of SDPs (p<0.05; see Methods). Pairs consisting of probesets located close to one another, which correspondingly have nearby peaks of linkage, are disproportionately represented among significantly correlated pairs as indicated by the red diagonal.
Figure 2. Scatter plots showing pairs of _trans_-eQTL genes with significantly correlated expression profiles by transcript genetic map location.
Significantly correlated pairs of _trans_-eQTL genes identified in a) fat, b) kidney, c) adrenal and d) left ventricle are plotted according to genetic map location of the transcripts. Colour coding is as in Figure 1.
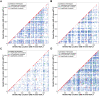
Figure 3. Scatter plots showing pairs of _trans_-eQTL genes with significantly correlated expression profiles by genetic map location of peak of linkage.
Significantly correlated pairs of _trans_-eQTLs identified in a) fat, b) kidney, c) adrenal and d) left ventricle are plotted according to genetic map position of the peak of linkage. Colour coding is as in Figure 1. Over-representation of pairs of _trans_-eQTLs with nearby peaks of linkage among the significantly correlated set can be observed (in the red diagonal) in all four tissues.
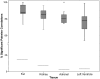
Figure 4. Boxplots showing percentage within-cluster correlation of genes underlying _trans_-eQTLs forming _trans_-eQTL clusters.
For each of the four tissues, the percentages of significantly correlated pairs of _trans_-eQTL genes within _trans_-eQTL clusters are displayed. The boxplots indicate the median, interquartile range, and range of within-cluster correlation in each tissue. The percentage significant correlation among all trans-eQTLs is shown for each of the four tissues as horizontal lines, for purpose of comparison with the _trans_-eQTL clusters. One outlier is shown, a cluster in adrenal found to have 100% significant pairwise correlation of gene expression levels (see Table S3).
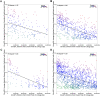
Figure 5. Scatter plots showing correlation of genes underlying cluster _trans_-eQTLs and _cis_-eQTLs located in the window region, plotted against their distance from the peak of linkage.
Each point on the scatter plot represents a _cis_-eQTL within the defined window region of a _trans_-eQTL cluster. The average Pearson coefficient of correlation of the underlying transcript expression levels of the _cis_-eQTL with those of the cluster _trans_-eQTLs is shown to be strongly negatively correlated with distance in a) fat, b) kidney, c) adrenal, d) LV, R2 for this relationship ranges from 0.15 to 0.43. The Z-score, calculated from the vertical distance of each _cis_-eQTL from the regression line, is indicated by the colour of each point, indicated by the colour legend in the top right of each plot.
Similar articles
- Mediation analysis demonstrates that trans-eQTLs are often explained by cis-mediation: a genome-wide analysis among 1,800 South Asians.
Pierce BL, Tong L, Chen LS, Rahaman R, Argos M, Jasmine F, Roy S, Paul-Brutus R, Westra HJ, Franke L, Esko T, Zaman R, Islam T, Rahman M, Baron JA, Kibriya MG, Ahsan H. Pierce BL, et al. PLoS Genet. 2014 Dec 4;10(12):e1004818. doi: 10.1371/journal.pgen.1004818. eCollection 2014 Dec. PLoS Genet. 2014. PMID: 25474530 Free PMC article. Clinical Trial. - Heritability and tissue specificity of expression quantitative trait loci.
Petretto E, Mangion J, Dickens NJ, Cook SA, Kumaran MK, Lu H, Fischer J, Maatz H, Kren V, Pravenec M, Hubner N, Aitman TJ. Petretto E, et al. PLoS Genet. 2006 Oct 20;2(10):e172. doi: 10.1371/journal.pgen.0020172. Epub 2006 Aug 28. PLoS Genet. 2006. PMID: 17054398 Free PMC article. - Causal network inference of cis- and trans-gene regulation of expression quantitative trait loci across human tissues.
Kvamme J, Badsha MB, Martin EA, Wu J, Wang X, Fu AQ. Kvamme J, et al. Genetics. 2025 Jun 4;230(2):iyaf064. doi: 10.1093/genetics/iyaf064. Genetics. 2025. PMID: 40179003 - Revealing the architecture of gene regulation: the promise of eQTL studies.
Gilad Y, Rifkin SA, Pritchard JK. Gilad Y, et al. Trends Genet. 2008 Aug;24(8):408-15. doi: 10.1016/j.tig.2008.06.001. Epub 2008 Jul 1. Trends Genet. 2008. PMID: 18597885 Free PMC article. Review. - Expression Quantitative Trait Loci Information Improves Predictive Modeling of Disease Relevance of Non-Coding Genetic Variation.
Croteau-Chonka DC, Rogers AJ, Raj T, McGeachie MJ, Qiu W, Ziniti JP, Stubbs BJ, Liang L, Martinez FD, Strunk RC, Lemanske RF Jr, Liu AH, Stranger BE, Carey VJ, Raby BA. Croteau-Chonka DC, et al. PLoS One. 2015 Oct 16;10(10):e0140758. doi: 10.1371/journal.pone.0140758. eCollection 2015. PLoS One. 2015. PMID: 26474488 Free PMC article. Review.
Cited by
- The gene balance hypothesis: implications for gene regulation, quantitative traits and evolution.
Birchler JA, Veitia RA. Birchler JA, et al. New Phytol. 2010 Apr;186(1):54-62. doi: 10.1111/j.1469-8137.2009.03087.x. Epub 2009 Nov 19. New Phytol. 2010. PMID: 19925558 Free PMC article. Review. - Genome-Wide Detection of Gene Coexpression Domains Showing Linkage to Regions Enriched with Polymorphic Retrotransposons in Recombinant Inbred Mouse Strains.
Scott-Boyer MP, Deschepper CF. Scott-Boyer MP, et al. G3 (Bethesda). 2013 Apr 9;3(4):597-605. doi: 10.1534/g3.113.005546. G3 (Bethesda). 2013. PMID: 23550129 Free PMC article. - Small RNA expression and strain specificity in the rat.
Linsen SE, de Wit E, de Bruijn E, Cuppen E. Linsen SE, et al. BMC Genomics. 2010 Apr 19;11:249. doi: 10.1186/1471-2164-11-249. BMC Genomics. 2010. PMID: 20403161 Free PMC article. - Mapping eQTLs in the Norfolk Island genetic isolate identifies candidate genes for CVD risk traits.
Benton MC, Lea RA, Macartney-Coxson D, Carless MA, Göring HH, Bellis C, Hanna M, Eccles D, Chambers GK, Curran JE, Harper JL, Blangero J, Griffiths LR. Benton MC, et al. Am J Hum Genet. 2013 Dec 5;93(6):1087-99. doi: 10.1016/j.ajhg.2013.11.004. Am J Hum Genet. 2013. PMID: 24314549 Free PMC article. - Integrated genomic approaches to identification of candidate genes underlying metabolic and cardiovascular phenotypes in the spontaneously hypertensive rat.
Morrissey C, Grieve IC, Heinig M, Atanur S, Petretto E, Pravenec M, Hubner N, Aitman TJ. Morrissey C, et al. Physiol Genomics. 2011 Nov 7;43(21):1207-18. doi: 10.1152/physiolgenomics.00210.2010. Epub 2011 Aug 16. Physiol Genomics. 2011. PMID: 21846806 Free PMC article.
References
- Jansen RC, Nap J-P. Genetical genomics: the added value from segregation. Trends in Genetics. 2001;17:388–391. - PubMed
- Mehrabian M, Allayee H, Stockton J, Lum PY, Drake TA, et al. Integrating genotypic and expression data in a segregating mouse population to identify 5-lipoxygenase as a susceptibility gene for obesity and bone traits. Nat Genet. 2005;37:1224–1233. - PubMed
- Hubner N, Wallace CA, Zimdahl H, Petretto E, Schulz H, et al. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat Genet. 2005;37:243–253. - PubMed
Publication types
MeSH terms
Grants and funding
- BHF_/British Heart Foundation/United Kingdom
- 069962/Z/02/Z/WT_/Wellcome Trust/United Kingdom
- MC_U120097112/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- MC_U120085815/MRC_/Medical Research Council/United Kingdom
- MC_U120061454/MRC_/Medical Research Council/United Kingdom
- HHMI/Howard Hughes Medical Institute/United States
- DH_/Department of Health/United Kingdom