Reaction of AdoMet with ThiC generates a backbone free radical - PubMed (original) (raw)
Reaction of AdoMet with ThiC generates a backbone free radical
N Cecilia Martinez-Gomez et al. Biochemistry. 2009.
Abstract
ThiC is an [4Fe-4S] cluster protein that catalyzes the formation of 4-amino-5-hydroxymethyl-2-methylpyrimidine. EPR spectroscopic studies demonstrate that, upon interaction with AdoMet, active ThiC from Salmonella enterica generates a persistent free radical on the alpha-carbon of an amino acid residue. The EPR properties of the radical are consistent with any residue other than a Gly or Ala. Exposure to oxygen was accompanied by a fission of the radical-carrying polypeptide chain between the Gly436 and His437 residues in ThiC. Regardless of whether the backbone radical is part of the catalytic machinery, its presence provides evidence that ThiC employs free radical chemistry as expected for radical SAM enzymes.
Figures
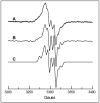
Figure 1
EPR spectrum of ThiC backbone radical. A) EPR spectrum at 40 K of reconstituted ThiC in the presence of AdoMet. The spectrum is an average of 4 scans with microwave power 4 mW, modulation amplitude 4 Gauss. B) Resolution enhanced version of spectrum A. C) Simulation of B assuming coupling to two β-protons (Aiso H1 = 15 G, Aiso H2 = 0.2 G); one β-amide 14N (A14N [0 G, 15 G, 0 G]; and one β-amide proton (A1H [8.4 G, 7.4 G, 0.2 G]. Additional details regarding the fitting parameters are given in supporting information.
Figure 2
Oxygen Fragmentation of the polypeptide chain. Reduced ThiC+AdoMet gassed with O2 for 20 minutes or kept under anoxic conditions as indicated. The samples were resolved by SDS-PAGE. A) for the high molecular weight bands (4-12%), and B) for the low molecular weight bands (12%). 2 μg protein was applied per lane, visualization was by Commassie blue staining. The sequence of ThiC surrounding the cleavage site is depicted. Starred residues are conserved in over 100 ThiC homologs. The line between Gly436 and His437 indicates the site of oxygen dependent cleavage.
Similar articles
- Structural diversity in the AdoMet radical enzyme superfamily.
Dowling DP, Vey JL, Croft AK, Drennan CL. Dowling DP, et al. Biochim Biophys Acta. 2012 Nov;1824(11):1178-95. doi: 10.1016/j.bbapap.2012.04.006. Epub 2012 Apr 28. Biochim Biophys Acta. 2012. PMID: 22579873 Free PMC article. Review. - S-Adenosylmethionine-dependent reduction of lysine 2,3-aminomutase and observation of the catalytically functional iron-sulfur centers by electron paramagnetic resonance.
Lieder KW, Booker S, Ruzicka FJ, Beinert H, Reed GH, Frey PA. Lieder KW, et al. Biochemistry. 1998 Feb 24;37(8):2578-85. doi: 10.1021/bi972417w. Biochemistry. 1998. PMID: 9485408 - Electron-nuclear double resonance spectroscopic evidence that S-adenosylmethionine binds in contact with the catalytically active 4Fe-4S cluster of pyruvate formate-lyase activating enzyme.
Walsby CJ, Hong W, Broderick WE, Cheek J, Ortillo D, Broderick JB, Hoffman BM. Walsby CJ, et al. J Am Chem Soc. 2002 Mar 27;124(12):3143-51. doi: 10.1021/ja012034s. J Am Chem Soc. 2002. PMID: 11902903 - Paramagnetic intermediates generated by radical S-adenosylmethionine (SAM) enzymes.
Stich TA, Myers WK, Britt RD. Stich TA, et al. Acc Chem Res. 2014 Aug 19;47(8):2235-43. doi: 10.1021/ar400235n. Epub 2014 Jul 3. Acc Chem Res. 2014. PMID: 24991701 Free PMC article. - Identification and function of auxiliary iron-sulfur clusters in radical SAM enzymes.
Lanz ND, Booker SJ. Lanz ND, et al. Biochim Biophys Acta. 2012 Nov;1824(11):1196-212. doi: 10.1016/j.bbapap.2012.07.009. Epub 2012 Jul 28. Biochim Biophys Acta. 2012. PMID: 22846545 Review.
Cited by
- Salmonella enterica synthesizes 5,6-dimethylbenzimidazolyl-(DMB)-α-riboside. Why some Firmicutes do not require the canonical DMB activation system to synthesize adenosylcobalamin.
Mattes TA, Escalante-Semerena JC. Mattes TA, et al. Mol Microbiol. 2017 Jan;103(2):269-281. doi: 10.1111/mmi.13555. Epub 2016 Nov 22. Mol Microbiol. 2017. PMID: 27748967 Free PMC article. - Radical S-adenosylmethionine enzymes.
Broderick JB, Duffus BR, Duschene KS, Shepard EM. Broderick JB, et al. Chem Rev. 2014 Apr 23;114(8):4229-317. doi: 10.1021/cr4004709. Epub 2014 Jan 29. Chem Rev. 2014. PMID: 24476342 Free PMC article. Review. No abstract available. - Characterization and mechanistic studies of DesII: a radical S-adenosyl-L-methionine enzyme involved in the biosynthesis of TDP-D-desosamine.
Szu PH, Ruszczycky MW, Choi SH, Yan F, Liu HW. Szu PH, et al. J Am Chem Soc. 2009 Oct 7;131(39):14030-42. doi: 10.1021/ja903354k. J Am Chem Soc. 2009. PMID: 19746907 Free PMC article. - High-resolution crystal structure of the eukaryotic HMP-P synthase (THIC) from Arabidopsis thaliana.
Coquille S, Roux C, Mehta A, Begley TP, Fitzpatrick TB, Thore S. Coquille S, et al. J Struct Biol. 2013 Dec;184(3):438-44. doi: 10.1016/j.jsb.2013.10.005. Epub 2013 Oct 23. J Struct Biol. 2013. PMID: 24161603 Free PMC article. - Mechanistic studies of the radical S-adenosyl-L-methionine enzyme DesII: EPR characterization of a radical intermediate generated during its catalyzed dehydrogenation of TDP-D-quinovose.
Ruszczycky MW, Choi SH, Mansoorabadi SO, Liu HW. Ruszczycky MW, et al. J Am Chem Soc. 2011 May 18;133(19):7292-5. doi: 10.1021/ja201212f. Epub 2011 Apr 22. J Am Chem Soc. 2011. PMID: 21513273 Free PMC article.
References
- Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. - PMC - PubMed
- Dougherty MJ, Downs DM. A connection between iron-sulfur cluster metabolism and the biosynthesis of 4-amino-5-hydroxymethyl-2-methylpyrimidine pyrophosphate in Salmonella enterica. Microbiol. 2006;152:2345–2353. - PubMed
- Wang SC, Frey PA. S-adenosylmethionine as an oxidant: the radical SAM superfamily. Trends Biochem Sci. 2007;32:101–110. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM047296/GM/NIGMS NIH HHS/United States
- R01 GM047296-16/GM/NIGMS NIH HHS/United States
- F32 GM082085/GM/NIGMS NIH HHS/United States
- F32 GM082085-02/GM/NIGMS NIH HHS/United States
- R56 GM035752/GM/NIGMS NIH HHS/United States
- R56 GM035752-22/GM/NIGMS NIH HHS/United States
- GM47296/GM/NIGMS NIH HHS/United States
- R56-GM35752/GM/NIGMS NIH HHS/United States
- GM 082085/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases