DGCR8-dependent microRNA biogenesis is essential for skin development - PubMed (original) (raw)
DGCR8-dependent microRNA biogenesis is essential for skin development
Rui Yi et al. Proc Natl Acad Sci U S A. 2009.
Abstract
MicroRNAs play important roles in animal development. Numerous conditional knockout (cKO) studies of Dicer have been performed to interrogate the functions of microRNA during mammalian development. However, because Dicer was recently implicated in the biogenesis of endogenous siRNAs in mammals, it raises the question whether the Dicer cKO defects can be attributable to the loss of microRNAs. Previously, we and others conditionally targeted Dicer and identified its critical roles in embryonic skin morphogenesis. Here, we focus explicitly on microRNAs by taking a parallel strategy with Dgcr8, encoding an essential component of the microprocessor complex that is exclusively required for microRNA biogenesis. With this comparative analysis, we show definitively that the Dicer- and Dgcr8-null skin defects are both striking and indistinguishable. By deep sequencing analysis of microRNA depletion in both Dicer- and Dgcr8-null skin, we demonstrate that most abundantly expressed skin microRNAs are dependent on both Dicer and DGCR8. Our results underscore a specific importance of microRNAs in controlling mammalian skin development.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Strategy for generating the Dgcr8 floxed allele in mouse genomic DNA (gDNA) and verification that microRNAs were effectively depleted as a result of Dgcr8 skin-specific ablation in mice. (A) Exon 3 within the Dgcr8 coding region was chosen for targeting because its deletion was predicted to generate a frameshift mutation and early truncation of the DGCR8 protein. Arrowheads denote positioning of the Lox sequences, recognized by Cre recombinase. WW, a WW protein–protein interaction module; dsRBD, double-stranded RNA-binding domain. (B) Conditional (_K14_-Cre) ablation of both Dgcr8 and Dicer in skin resulted in the depletion of microRNAs compared with their WT counterparts. The expression levels of three representative microRNAs highly expressed in skin were chosen for analyses. Values for WT samples were designated as 1. SnoRNA25 served as the internal control.
Fig. 2.
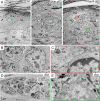
Skin phenotypes of Dgcr8 and Dicer cKO mice are strikingly similar. (A) Dgcr8 and Dicer cKO mice can survive up to 5–6 days after birth. Newborn (P0.5) cKO and WT mice are similar in size and appearance. Thereafter, cKO mice fail to gain weight and exhibit taught, flaky skin, a sign of severe dehydration. (B) P6 Dgcr8 and Dicer cKO skins display evaginating hair germs (hg) that appear as balls of undifferentiated cells (arrows) that distort the epidermis (epi). (Scale bar, 10 μm.) (C) Immunofluorescence identifies germs molecularly by transcription factor Lef1 and by hemidesmosomal β4 integrin (nonspecific dermal staining is caused by the secondary antibody). (Scale bar, 20 μm.) (D) Active caspase-3 (Cas3) denotes significant enrichment of apoptotic cells within hair germs in the cKO skin. Dotted lines mark epidermal-dermal boundaries. der, dermis. (Scale bar, 20 μm.)
Fig. 3.
Ultrastructural defects of Dicer and Dgcr8 cKO skin. (A) Skins from neonatal WT control, Dicer cKO and Dgcr8 cKO mice were fixed and processed for electron microscopy as described. Shown are regions of the epidermis, where a whorl of evaginated hair germ (hg) cells are readily identified by the presence of dermal papilla (DP) cells at the center of the structure. These aberrations in follicle morphogenesis distorted the surrounding epidermis. (Scale bar, 10 μm.) (B) Cells with hair follicle matrix cell (Mx) morphology were present in the evaginating hair germs of Dgcr8 cKO epidermis. These cells frequently contained melanin granules (Mel). Melanocytes are normally never in mouse skin epidermis, and the appearance of melanin within the whorls was an additional hallmark of the evaginating hair germs. (Scale bar, 2 μm.) (C and E) Closed-up pictures of boxed areas in A. Note the presence of hemidesmosomes (Hd) and a loose basal lamina (bl) at the evaginating hair germ-DP border (C) as well as the epidermal-dermal boundary to what appeared to be an underlying DP (E) in the Dgcr8 cKO skin. (Scale bar, 500 nm.) (D) Enriched apoptotic cells (Ap) in hair germs of Dgcr8 cKO skin. (Scale bar, 2 μm.) Additional abbreviations: SC, stratum corneum; Gr, granular layers; Sp, spinous layers; BL, basal epidermal layer; Der, dermis. Dotted lines mark epidermal-dermal boundaries.
Fig. 4.
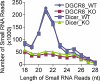
Specific and similar depletion of small RNA reads (19–23 nt) in both Dicer and Dgcr8 cKO skin. Small RNA reads from 18 to 28 nucleotides were charted from all four small RNA cDNA libraries. Note the nearly identical distribution patterns of small RNA reads within either two duplicate WT libraries or two cKO libraries.
Fig. 5.
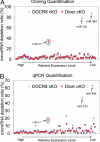
Small RNA cloning and deep sequencing reveals the dependence of microRNA biogenesis on Dicer and DGCR8. (A) Compilation of microRNA sequencing data in Dicer and Dgcr8 cKO samples compared with their WT littermate skins reveal similar dependence of microRNA biogenesis with the exception for miR-320 and miR-484, which require Dicer but not DGCR8. MiR-21 was depleted in both Dicer and Dgcr8 cKO, but only partially (≈70%, circle). MicroRNA depletion ratios were normalized against reads from rRNA, tRNA, and spiked-in calibration RNAs, none of which depends on Dicer or DGCR8. (B) qPCR microRNA expression validation of the cloning/sequencing results. Of 67 skin microRNAs subjected to quantification by qPCR, 65 were dependent similarly on Dicer and DGCR8. MiR-320 and miR-484 displayed dependency on Dicer but not DGCR8.
Similar articles
- Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs.
Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Babiarz JE, et al. Genes Dev. 2008 Oct 15;22(20):2773-85. doi: 10.1101/gad.1705308. Genes Dev. 2008. PMID: 18923076 Free PMC article. - MicroRNA function is globally suppressed in mouse oocytes and early embryos.
Suh N, Baehner L, Moltzahn F, Melton C, Shenoy A, Chen J, Blelloch R. Suh N, et al. Curr Biol. 2010 Feb 9;20(3):271-7. doi: 10.1016/j.cub.2009.12.044. Epub 2010 Jan 28. Curr Biol. 2010. PMID: 20116247 Free PMC article. - DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal.
Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. Wang Y, et al. Nat Genet. 2007 Mar;39(3):380-5. doi: 10.1038/ng1969. Epub 2007 Jan 28. Nat Genet. 2007. PMID: 17259983 Free PMC article. - Dgcr8 knockout approaches to understand microRNA functions in vitro and in vivo.
Guo WT, Wang Y. Guo WT, et al. Cell Mol Life Sci. 2019 May;76(9):1697-1711. doi: 10.1007/s00018-019-03020-9. Epub 2019 Jan 29. Cell Mol Life Sci. 2019. PMID: 30694346 Free PMC article. Review. - Dicing with viruses: microRNAs as antiviral factors.
Müller S, Imler JL. Müller S, et al. Immunity. 2007 Jul;27(1):1-3. doi: 10.1016/j.immuni.2007.07.003. Immunity. 2007. PMID: 17663977 Review.
Cited by
- Dendrite intercalation between epidermal cells tunes nociceptor sensitivity to mechanical stimuli in Drosophila larvae.
Luedke KP, Yoshino J, Yin C, Jiang N, Huang JM, Huynh K, Parrish JZ. Luedke KP, et al. PLoS Genet. 2024 Apr 25;20(4):e1011237. doi: 10.1371/journal.pgen.1011237. eCollection 2024 Apr. PLoS Genet. 2024. PMID: 38662763 Free PMC article. - Research progress and perspectives of noncoding RNAs in adrenocortical carcinoma: A review.
Xu C, Xu P, Zhang J, He S, Hua T, Huang A. Xu C, et al. Medicine (Baltimore). 2024 Jan 26;103(4):e36908. doi: 10.1097/MD.0000000000036908. Medicine (Baltimore). 2024. PMID: 38277554 Free PMC article. Review. - Identification of Potential miRNA-mRNA Regulatory Network Associated with Growth and Development of Hair Follicles in Forest Musk Deer.
Qi WH, Liu T, Zheng CL, Zhao Q, Zhou N, Zhao GJ. Qi WH, et al. Animals (Basel). 2023 Dec 15;13(24):3869. doi: 10.3390/ani13243869. Animals (Basel). 2023. PMID: 38136906 Free PMC article. - Adipose-derived stem cell exosome NFIC improves diabetic foot ulcers by regulating miR-204-3p/HIPK2.
Huang H, Zhu W, Huang Z, Zhao D, Cao L, Gao X. Huang H, et al. J Orthop Surg Res. 2023 Sep 14;18(1):687. doi: 10.1186/s13018-023-04165-x. J Orthop Surg Res. 2023. PMID: 37710299 Free PMC article. - Identification and functional validation of SRC and RAPGEF1 as new direct targets of miR-203, involved in regulation of epidermal homeostasis.
Golebiewski C, Gastaldi C, Vieu DL, Mari B, Rezzonico R, Bernerd F, Marionnet C. Golebiewski C, et al. Sci Rep. 2023 Aug 27;13(1):14006. doi: 10.1038/s41598-023-40441-w. Sci Rep. 2023. PMID: 37635193 Free PMC article.
References
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. - PubMed
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. - PubMed
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K99 AR054704/AR/NIAMS NIH HHS/United States
- R00 AR054704/AR/NIAMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- K99AR054704/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases