Identification of direct residue contacts in protein-protein interaction by message passing - PubMed (original) (raw)
Identification of direct residue contacts in protein-protein interaction by message passing
Martin Weigt et al. Proc Natl Acad Sci U S A. 2009.
Abstract
Understanding the molecular determinants of specificity in protein-protein interaction is an outstanding challenge of postgenome biology. The availability of large protein databases generated from sequences of hundreds of bacterial genomes enables various statistical approaches to this problem. In this context covariance-based methods have been used to identify correlation between amino acid positions in interacting proteins. However, these methods have an important shortcoming, in that they cannot distinguish between directly and indirectly correlated residues. We developed a method that combines covariance analysis with global inference analysis, adopted from use in statistical physics. Applied to a set of >2,500 representatives of the bacterial two-component signal transduction system, the combination of covariance with global inference successfully and robustly identified residue pairs that are proximal in space without resorting to ad hoc tuning parameters, both for heterointeractions between sensor kinase (SK) and response regulator (RR) proteins and for homointeractions between RR proteins. The spectacular success of this approach illustrates the effectiveness of the global inference approach in identifying direct interaction based on sequence information alone. We expect this method to be applicable soon to interaction surfaces between proteins present in only 1 copy per genome as the number of sequenced genomes continues to expand. Use of this method could significantly increase the potential targets for therapeutic intervention, shed light on the mechanism of protein-protein interaction, and establish the foundation for the accurate prediction of interacting protein partners.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
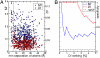
The combined covariance/message-passing approach detects 2 groups of correlated pairs. (A) Scatter plot of direct mutual information (DI) versus total mutual information (MI) reveals 2 classes of covarying residue pairs, those with strong direct correlation found in the upper red quadrant (group I) and those with low direct correlation found in the lower green quadrant (group II). A group of pairings just around the border of the MI and/or the DI cutoff is highlighted in blue. (B) Direct and indirect interaction pairs depicted on exemplary structures of the HisKA and RR domain. All residues that appear in the network of pairings with MI > MI(t) were mapped onto the structures of HK853 from T. maritima (HisKA domain) and Spo0F from B. subtilis (RR domain). Those pairings showing strong direct correlation are depicted in red and connected by a red line and those that show low direct correlation are depicted in green and connected by a green line. Green lines connecting red residues represent low direct correlation for that particular residue pairing. For orientation, the N and C termini and relevant structural elements are labeled. The phosphotransfer sites H260 in HK853 and D54 in Spo0F are shown in yellow.
Fig. 2.
Direct Information is inversely correlated with residue distance of pairs in the Spo0B/Spo0F cocrystal structure. (A) Minimal atom distance for all 408 pairings that could be mapped to the Spo0B/Spo0F cocrystal structure was determined in Ångström and plotted either against direct information DI (red symbols) or total mutual information MI (blue symbols). (B) Specificity vs. rank percentile for predicting contact pairs via DI (red curve) and MI (blue curve). Specificity is defined as the fraction of pairings at the given rank percentile that are within 6Å in the Spo0B/Spo0F cocrystal structure.
Fig. 3.
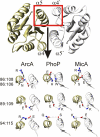
Direct interaction between the identified dimer contact pairs. Four dimer contact pairings (red entries in
Table S1
) are localized to the α4- and α5-helices and are shown on the exemplary OmpR class RR structures of ArcA, PhoP, and MicA as a dashed line. Whereas contact pairing 89:109 (ArcA numbering, for PhoP and MicA numbering deduct 2) happens to represent a salt bridge in all 3 structural examples shown here, the pairing 94:115 and the cluster involving pairings 86:106 and 86:108 demonstrate nicely the type of residue variation that is the basis of the covariance method (see text). Detailed analysis of the covariance among the residues involved in these 4 pairings are given in
Fig. S8
.
Similar articles
- Inference of direct residue contacts in two-component signaling.
Lunt B, Szurmant H, Procaccini A, Hoch JA, Hwa T, Weigt M. Lunt B, et al. Methods Enzymol. 2010;471:17-41. doi: 10.1016/S0076-6879(10)71002-8. Epub 2010 Mar 1. Methods Enzymol. 2010. PMID: 20946840 - Accurate prediction of protein-protein interactions from sequence alignments using a Bayesian method.
Burger L, van Nimwegen E. Burger L, et al. Mol Syst Biol. 2008;4:165. doi: 10.1038/msb4100203. Epub 2008 Feb 12. Mol Syst Biol. 2008. PMID: 18277381 Free PMC article. - Interaction fidelity in two-component signaling.
Szurmant H, Hoch JA. Szurmant H, et al. Curr Opin Microbiol. 2010 Apr;13(2):190-7. doi: 10.1016/j.mib.2010.01.007. Epub 2010 Feb 3. Curr Opin Microbiol. 2010. PMID: 20133181 Free PMC article. Review. - Computational prediction of protein-protein interactions.
Obenauer JC, Yaffe MB. Obenauer JC, et al. Methods Mol Biol. 2004;261:445-68. doi: 10.1385/1-59259-762-9:445. Methods Mol Biol. 2004. PMID: 15064475 Review.
Cited by
- Evolutionary history expands the range of signaling interactions in hybrid multikinase networks.
Ortet P, Fochesato S, Bitbol AF, Whitworth DE, Lalaouna D, Santaella C, Heulin T, Achouak W, Barakat M. Ortet P, et al. Sci Rep. 2021 Jun 3;11(1):11763. doi: 10.1038/s41598-021-91260-w. Sci Rep. 2021. PMID: 34083699 Free PMC article. - Mi3-GPU: MCMC-based Inverse Ising Inference on GPUs for protein covariation analysis.
Haldane A, Levy RM. Haldane A, et al. Comput Phys Commun. 2021 Mar;260:107312. doi: 10.1016/j.cpc.2020.107312. Epub 2020 Apr 17. Comput Phys Commun. 2021. PMID: 33716309 Free PMC article. - Inferring interaction partners from protein sequences.
Bitbol AF, Dwyer RS, Colwell LJ, Wingreen NS. Bitbol AF, et al. Proc Natl Acad Sci U S A. 2016 Oct 25;113(43):12180-12185. doi: 10.1073/pnas.1606762113. Epub 2016 Sep 23. Proc Natl Acad Sci U S A. 2016. PMID: 27663738 Free PMC article. - Protein structure prediction from sequence variation.
Marks DS, Hopf TA, Sander C. Marks DS, et al. Nat Biotechnol. 2012 Nov;30(11):1072-80. doi: 10.1038/nbt.2419. Nat Biotechnol. 2012. PMID: 23138306 Free PMC article. - Comparative study of the effectiveness and limitations of current methods for detecting sequence coevolution.
Mao W, Kaya C, Dutta A, Horovitz A, Bahar I. Mao W, et al. Bioinformatics. 2015 Jun 15;31(12):1929-37. doi: 10.1093/bioinformatics/btv103. Epub 2015 Feb 19. Bioinformatics. 2015. PMID: 25697822 Free PMC article.
References
- Cusick ME, Klitgord N, Vidal M, Hill DE. Interactome: Gateway into systems biology. Hum Mol Genet. 2005;14((Spec No 2)):R171–R181. - PubMed
- Kortemme T, Baker D. Computational design of protein-protein interactions. Curr Opin Chem Biol. 2004;8:91–97. - PubMed
- Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. - PubMed
- Suel GM, Lockless SW, Wall MA, Ranganathan R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol. 2003;10:59–69. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources