Regulation of estrogen-dependent transcription by the LIM cofactors CLIM and RLIM in breast cancer - PubMed (original) (raw)
. 2009 Jan 1;69(1):128-36.
doi: 10.1158/0008-5472.CAN-08-1630.
Cenap Güngör, Tanja Prenzel, Sabine Riethdorf, Lutz Riethdorf, Naoko Taniguchi-Ishigaki, Thomas Rau, Baris Tursun, J David Furlow, Guido Sauter, Martin Scheffner, Klaus Pantel, Frank Gannon, Ingolf Bach
Affiliations
- PMID: 19117995
- PMCID: PMC2713826
- DOI: 10.1158/0008-5472.CAN-08-1630
Regulation of estrogen-dependent transcription by the LIM cofactors CLIM and RLIM in breast cancer
Steven A Johnsen et al. Cancer Res. 2009.
Abstract
Mammary oncogenesis is profoundly influenced by signaling pathways controlled by estrogen receptor alpha (ERalpha). Although it is known that ERalpha exerts its oncogenic effect by stimulating the proliferation of many human breast cancers through the activation of target genes, our knowledge of the underlying transcriptional mechanisms remains limited. Our published work has shown that the in vivo activity of LIM homeodomain transcription factors (LIM-HD) is critically regulated by cofactors of LIM-HD proteins (CLIM) and the ubiquitin ligase RING finger LIM domain-interacting protein (RLIM). Here, we identify CLIM and RLIM as novel ERalpha cofactors that colocalize and interact with ERalpha in primary human breast tumors. We show that both cofactors associate with estrogen-responsive promoters and regulate the expression of endogenous ERalpha target genes in breast cancer cells. Surprisingly, our results indicate opposing functions of LIM cofactors for ERalpha and LIM-HDs: whereas CLIM enhances transcriptional activity of LIM-HDs, it inhibits transcriptional activation mediated by ERalpha on most target genes in vivo. In turn, the ubiquitin ligase RLIM inhibits transcriptional activity of LIM-HDs but enhances transcriptional activation of endogenous ERalpha target genes. Results from a human breast cancer tissue microarray of 1,335 patients revealed a highly significant correlation of elevated CLIM levels to ER/progesterone receptor positivity and poor differentiation of tumors. Combined, these results indicate that LIM cofactors CLIM and RLIM regulate the biological activity of ERalpha during the development of human breast cancer.
Figures
Figure 1
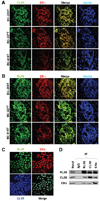
CLIM and RLIM are co-localized with ERα in the nucleus of breast cancer cells. Immunohistochemical staining of breast tumor samples was performed on cryosections using an anti-ERα monoclonal antibody together with anti-CLIM (A) or anti-RLIM (B) polyclonal antibodies. CLIM and RLIM expression is observed almost exclusively in the nucleus and is co-localized with ERα expression. CLIM, RLIM and ERα are also co-localized in the nucleus of MCF7 breast cancer cells (C). (D) Interaction of endogenous RLIM, CLIM and ERα in MCF7 cells. Total cell extracts of MCF7 cells were immunoprecipitated with specific anti-RLIM, -CLIM, -ERα or non-specific IgG polyclonal antibodies. Western blot analysis was performed with each antibody. Twenty percent input is shown.
Figure 2
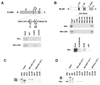
CLIM and RLIM are specific ERα-interacting proteins. 35S-labeled in vitro transcribed and translated full-length human ERα (A–D), isolated human ERα-LBD (A,B), full-length human ERα ERβ (C), full-length human AR (D) and full-length human ERα GR (D) were tested for their ability to interact with full-length GST-CLIM2 or GST-RLIM (A–D) or various mutants of CLIM2 (A) or RLIM (B). Equal amounts of GST and GST-fusion proteins were used and input samples of in vitro translated proteins were loaded separately. (A) CLIM contains a RID (upper panel) within the LCCD domain which is necessary for interaction with full-length (middle panel) ERα and with the isolated ERα-LBD (lower panel). DD, dimerization domain; LCCD, LDB/Chip conserved domain; NLS, nuclear localization signal; LID, LIM-interacting domain. (B) RLIM interacts with the LBD of ERα through the C-terminal RING finger-containing domain. NLS, nuclear localization signal; BD, basic domain; RING, RING-H2 zinc finger. CLIM2 and RLIM specifically interact with ERα, but not with ERβ (C), AR (D) or GR (D) in the presence of the respective ligands.
Figure 3
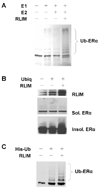
RLIM targets ERα for ubiquitination. (A) ERα is an in vitro substrate for ubiquitination by RLIM. In vitro 35S-labelled full-length ERα protein was incubated alone or together with various combinations of the ubiquitin activating enzyme (E1), the ubiquitin conjugating enzyme (E2) UbcH5 or bacterially expressed and purified full-length GST-RLIM. The brackets indicate higher molecular weight ubiquitinated ERα. Ub-ERα, ubiquitinated ERα. (B) Overexpression of RLIM increases the formation of high molecular weight, detergent insoluble forms of ERα. H1299 cells were transfected with ERα and ubiquitin expression vectors as indicated. Whole extracts were prepared and detergent soluble and insoluble fractions were separately analyzed by western blot analysis with specific anti-RLIM polyclonal or anti-ERα monoclonal antibodies. Note that there is a noticeable increase in a high molecular weight form of ERα upon overexpression of RLIM in the detergent insoluble fraction which is not apparent in the detergent soluble fraction. Ubiq, ubiquitin. (C) Overexpression of RLIM results in increased ERα ubiquitination in cells. H1299 cells were transfected with a His-tagged ubiquitin (His-Ub) expression vector with or without RLIM overexpression as in (B) and the ubiquitinated proteins were isolated using a Ni-NTA affinity matrix and analyzed by western blot using a specific anti-ERα monoclonal antibody. The brackets indicate the increase in high molecular weight ubiquitinated ERα upon RLIM overexpression.
Figure 4
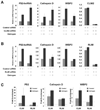
CLIM and RLIM regulate the transcriptional activity of ERα. (A) CLIM overexpression dose-dependently decreases ERE transcriptional activity. MCF7 cells were transfected with an ERE-containing luciferase reporter construct (ERE-TK-Luc) and an internal control plasmid for transfection efficiency (phRG-TK) which constitutively expresses Renilla luciferase together with the indicated amounts of a plasmid expressing CLIM2 or a CLIM2 RID mutant (AxxAA). The total amount of DNA was kept constant by adding an appropriate amount of control plasmid (pCS2). Estrogen (10−8 M ethinyl estradiol) was added 24h after transfection and cells were harvested after another 24h. All transfections were normalized and expressed relative to the average of the estrogen-treated, control transfected cells as relative activity. (B) RLIM overexpression dose-dependently increases ERE transcriptional activity. MCF7 cells were transfected with ERE-TK-Luc and phRG-TK together with the indicated amounts of a RLIM or RLIMΔRING expression vector and treated with estrogen as in (A). The total amount of plasmid DNA was kept constant by adding the control plasmid (pCS2). (C) CLIM2 and RLIM siRNA increase and decrease ERE activity, respectively. ERE-TK-Luc and phRG-TK were transfected as in (A) and (B) together with control, CLIM2 or RLIM siRNA. Cells were grown for 48h to allow for a knock-down of endogenous CLIM2 or RLIM prior to estrogen treatment for another 24h. Luciferase activity was expressed as relative activity as in (A) and (B).
Figure 5
CLIM2 and RLIM regulate endogenous estrogen receptor activity. (A) Endogenous CLIM2 regulates ERα activity in cells. MCF7 breast cancer cells were transfected with control siRNA (A and B), CLIM2 siRNA (A) or RLIM siRNA (B). Cells were grown for 48 hours and treated for 2h with estrogen (10−8 M ethinyl estradiol) or untreated. Gene expression was measured by quantitative RT-PCR using primers specific for PS2-hnRNA, Cathepsin D, WISP2 or CLIM2 mRNA. mRNA levels were normalized to an unregulated gene (36B4) and expressed relative to the uninduced control siRNA transfected cells. CLIM2 mRNA levels are shown to verify efficient knock-down. (B) RLIM is necessary for optimal induction of gene expression by the endogenous estrogen receptor. MCF7 cells were transfected with RLIM siRNA and treated as in (A). In opposition to CLIM2 siRNA, RLIM siRNA significantly decreases the induction of estrogen-regulated gene expression compared to the control conditions. RLIM mRNA levels are decreased approximately 90% upon siRNA transfection. (C) CLIM and RLIM are recruited to endogenous EREs. CLIM or RLIM recruitment to the PS2, Cathepsin D and WISP2 genes was analyzed by chromatin immunoprecipitation analysis using chromatin from MCF7 cells untreated or treated with estrogen (10−8 M ethinyl estradiol) for 1h. Estrogen treatment increased the recruitment of both CLIM and RLIM to the PS2 and WISP2 genes while only CLIM recruitment was also increased on the Cathepsin D gene where RLIM was also present in an estrogen-independent manner. Non-specific IgG was used to distinguish between specific and background binding. ChIP samples were normalized to input samples and expressed as fold enrichment relative to the average of all IgG ChIP samples.
Similar articles
- Ubiquitination-dependent cofactor exchange on LIM homeodomain transcription factors.
Ostendorff HP, Peirano RI, Peters MA, Schlüter A, Bossenz M, Scheffner M, Bach I. Ostendorff HP, et al. Nature. 2002 Mar 7;416(6876):99-103. doi: 10.1038/416099a. Nature. 2002. PMID: 11882901 - Dynamic expression of LIM cofactors in the developing mouse neural tube.
Ostendorff HP, Tursun B, Cornils K, Schlüter A, Drung A, Güngör C, Bach I. Ostendorff HP, et al. Dev Dyn. 2006 Mar;235(3):786-91. doi: 10.1002/dvdy.20669. Dev Dyn. 2006. PMID: 16395690 - Negative regulation of estrogen receptor alpha transactivation functions by LIM domain only 4 protein.
Singh RR, Barnes CJ, Talukder AH, Fuqua SA, Kumar R. Singh RR, et al. Cancer Res. 2005 Nov 15;65(22):10594-601. doi: 10.1158/0008-5472.CAN-05-2268. Cancer Res. 2005. PMID: 16288053 - Breast cancer cells: Modulation by melatonin and the ubiquitin-proteasome system--a review.
Vriend J, Reiter RJ. Vriend J, et al. Mol Cell Endocrinol. 2015 Dec 5;417:1-9. doi: 10.1016/j.mce.2015.09.001. Epub 2015 Sep 9. Mol Cell Endocrinol. 2015. PMID: 26363225 Review. - Mechanisms of estrogen receptor-α upregulation in breast cancers.
Miyoshi Y, Murase K, Saito M, Imamura M, Oh K. Miyoshi Y, et al. Med Mol Morphol. 2010 Dec;43(4):193-6. doi: 10.1007/s00795-010-0514-3. Epub 2011 Jan 26. Med Mol Morphol. 2010. PMID: 21267694 Review.
Cited by
- Cofactors of LIM Domains Associate with Estrogen Receptor α to Regulate the Expression of Noncoding RNA H19 and Corneal Epithelial Progenitor Cell Function.
Klein RH, Stephens DN, Ho H, Chen JK, Salmans ML, Wang W, Yu Z, Andersen B. Klein RH, et al. J Biol Chem. 2016 Jun 17;291(25):13271-85. doi: 10.1074/jbc.M115.709386. Epub 2016 Apr 29. J Biol Chem. 2016. PMID: 27129775 Free PMC article. - [LIM-domain binding protein 2 regulated by m6A modification inhibits lung adenocarcinoma cell proliferation in vitro].
Zhai D, Wang G, Li L, Jia X, Zheng G, Yin J. Zhai D, et al. Nan Fang Yi Ke Da Xue Xue Bao. 2021 Mar 25;41(3):329-335. doi: 10.12122/j.issn.1673-4254.2021.03.03. Nan Fang Yi Ke Da Xue Xue Bao. 2021. PMID: 33849822 Free PMC article. Chinese. - Regulation of LIM-domain-binding 1 protein expression by ubiquitination of Lys134.
Howard PW, Jue SF, Ransom DG, Maurer RA. Howard PW, et al. Biochem J. 2010 Jul 1;429(1):127-36. doi: 10.1042/BJ20091461. Biochem J. 2010. PMID: 20423330 Free PMC article. - Multivariate models from RNA-Seq SNVs yield candidate molecular targets for biomarker discovery: SNV-DA.
Paul MR, Levitt NP, Moore DE, Watson PM, Wilson RC, Denlinger CE, Watson DK, Anderson PE. Paul MR, et al. BMC Genomics. 2016 Mar 31;17:263. doi: 10.1186/s12864-016-2542-4. BMC Genomics. 2016. PMID: 27029813 Free PMC article. - Crystal structure of human LDB1 in complex with SSBP2.
Wang H, Kim J, Wang Z, Yan XX, Dean A, Xu W. Wang H, et al. Proc Natl Acad Sci U S A. 2020 Jan 14;117(2):1042-1048. doi: 10.1073/pnas.1914181117. Epub 2019 Dec 31. Proc Natl Acad Sci U S A. 2020. PMID: 31892537 Free PMC article.
References
- Osborne CK, McGuire WL. The use of steroid hormone receptors in the treatment of human breast cancer: a review. Bull Cancer. 1979;66:203–209. - PubMed
- Maynard PV, Blamey RW, Elston CW, Haybittle JL, Griffiths K. Estrogen receptor assay in primary breast cancer and early recurrence of the disease. Cancer Res. 1978;38:4292–4295. - PubMed
- Rich MA, Furmanski P, Brooks SC. Prognostic value of estrogen receptor determinations in patients with breast cancer. Cancer Res. 1978;38:4296–4298. - PubMed
- Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. - PubMed
- Holst F, Stahl PR, Ruiz C, et al. Estrogen receptor alpha (ESR1) gene amplification is frequent in breast cancer. Nat Genet. 2007;39:655–660. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 CA108324/CA/NCI NIH HHS/United States
- R01 CA131158/CA/NCI NIH HHS/United States
- R01 CA131158-01A1/CA/NCI NIH HHS/United States
- F32 CA 108324/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous