Linking integrin conformation to function - PubMed (original) (raw)
Review
Linking integrin conformation to function
Janet A Askari et al. J Cell Sci. 2009.
Abstract
Integrins are alphabeta heterodimeric adhesion receptors that relay signals bidirectionally across the plasma membrane between the extracellular matrix and cell-surface ligands, and cytoskeletal and signalling effectors. The physical and chemical signals that are controlled by integrins are essential for intercellular communication and underpin all aspects of metazoan existence. To mediate such diverse functions, integrins exhibit structural diversity, flexibility and dynamism. Conformational changes, as opposed to surface expression or clustering, are central to the regulation of receptor function. In recent years, there has been intense interest in determining the three-dimensional structure of integrins, and analysing the shape changes that underpin the interconversion between functional states. Considering the central importance of the integrin signalling nexus, it is perhaps no surprise that obtaining this information has been difficult, and the answers gained so far have been complicated. In this Commentary, we pose some of the key remaining questions that surround integrin structure-function relationships and review the evidence that supports the current models.
Figures
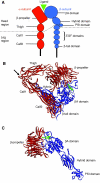
Fig. 1.
Integrin structure. (A) Schematic diagram of integrin structure. The overall structure is that of a head region [propeller and thigh domains of the α-subunit and the βA (also known as βI), hybrid and PSI domains of the β-subunit] supported on two legs that are made up of the calf1 and calf2 domains in the α-subunit and the EGF repeats and β-tail domain in the β-subunit. The binding of ligands takes place at an interface between the propeller domain and βA domain. (B) Ribbon diagram of the structure of the ectodomain of integrin αVβ3 in complex with the high-affinity ligand cyclic RGD peptide (Xiong et al., 2002). The α-subunit is shown in red, the β-subunit in blue; peptide is shown as a ball-and-stick model with atoms in green. Metal ions (silver spheres) occupy the base of the propeller and the top face of the βA domain. The protein is in a closed form, which is bent at the knees or `genu' (arrow). Some β-subunit domains are not visible in the structure. (C) Ribbon diagram of the structure of the head region of integrin αIIbβ3 in complex with the high-affinity ligand eptifibatide (Xiao et al., 2004). Colour coding is the same as in B. In this open structure the hybrid domain has swung outwards and the leg regions (not present) would be unbent so that the integrin is in an extended conformation, similar to that depicted in A.
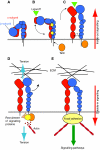
Fig. 2.
Integrin conformation-function relationships: a model. A five-component model illustrating conformational changes that are associated with inside-out and outside-in integrin signalling. The α-subunit is in red and the β-subunit in blue. The figure shows the three major conformational states that have been identified so far: inactive (A), primed (B) and ligand bound (C) (ligand is represented by a green triangle), together with possible intermediate conformers. Panels A-C represent conformations that mediate inside-out signalling, and panels D and E, outside-in signalling (the direction is indicated by red arrows). (A) Inactive integrin adopts a compact, most probably bent conformation in which the α- and β-subunit leg, transmembrane and cytoplasmic domains are closely associated. (B) The inherent flexibility of the knees allows for a degree of movement or `breathing' in this structure. Intracellular signals, culminating in the binding of talin (orange oval) to the β-subunit tail, causes relaxation of the leg restraints, allowing some further unbending that is sufficient to expose the epitopes of stimulatory antibodies in the leg regions (represented by yellow stars). A concomitant small outward movement of the hybrid domain primes the ligand-binding pocket to achieve a high-affinity conformation that is ready to accept ligand. The point at which a high-affinity conformation is reached may be integrin- and agonist-specific, and might take place before the receptor is fully extended. (C) The primed integrin binds ligand, which represents the end-point of inside-out signalling. At this stage the integrin is probably in an extended conformation, but the hybrid domain might remain in its primed position and, although some destabilisation and rearrangement of the legs has occurred, their degree of separation is not known. (D,E) The binding of talin and ligand initiate focal contact formation. As the cytoskeleton matures, tension (D, blue arrows) is generated on the integrin receptor across the cell membrane. (E) The force applied to the integrin headpiece triggers further outward movement of the hybrid domain, strengthening receptor-ligand binding and allowing the formation of stable focal adhesions and the initiation of intracellular signalling cascades (green arrow), the end-point of outside-in signalling.
Similar articles
- Migfilin, a molecular switch in regulation of integrin activation.
Ithychanda SS, Das M, Ma YQ, Ding K, Wang X, Gupta S, Wu C, Plow EF, Qin J. Ithychanda SS, et al. J Biol Chem. 2009 Feb 13;284(7):4713-22. doi: 10.1074/jbc.M807719200. Epub 2008 Dec 13. J Biol Chem. 2009. PMID: 19074766 Free PMC article. - The talin-tail interaction places integrin activation on FERM ground.
Campbell ID, Ginsberg MH. Campbell ID, et al. Trends Biochem Sci. 2004 Aug;29(8):429-35. doi: 10.1016/j.tibs.2004.06.005. Trends Biochem Sci. 2004. PMID: 15362227 Review. - A novel membrane-dependent on/off switch mechanism of talin FERM domain at sites of cell adhesion.
Song X, Yang J, Hirbawi J, Ye S, Perera HD, Goksoy E, Dwivedi P, Plow EF, Zhang R, Qin J. Song X, et al. Cell Res. 2012 Nov;22(11):1533-45. doi: 10.1038/cr.2012.97. Epub 2012 Jun 19. Cell Res. 2012. PMID: 22710802 Free PMC article. - Pull and push: talin activation for integrin signaling.
Wang JH. Wang JH. Cell Res. 2012 Nov;22(11):1512-4. doi: 10.1038/cr.2012.103. Epub 2012 Jul 10. Cell Res. 2012. PMID: 22777425 Free PMC article. - Structure and mechanics of integrin-based cell adhesion.
Arnaout MA, Goodman SL, Xiong JP. Arnaout MA, et al. Curr Opin Cell Biol. 2007 Oct;19(5):495-507. doi: 10.1016/j.ceb.2007.08.002. Epub 2007 Oct 24. Curr Opin Cell Biol. 2007. PMID: 17928215 Free PMC article. Review.
Cited by
- 18F-alfatide PET/CT may predict short-term outcome of concurrent chemoradiotherapy in patients with advanced non-small cell lung cancer.
Luan X, Huang Y, Gao S, Sun X, Wang S, Ma L, Teng X, Lu H, Yu J, Yuan S. Luan X, et al. Eur J Nucl Med Mol Imaging. 2016 Dec;43(13):2336-2342. doi: 10.1007/s00259-016-3505-3. Epub 2016 Sep 8. Eur J Nucl Med Mol Imaging. 2016. PMID: 27631310 Free PMC article. - Integrin-specific mechanoresponses to compression and extension probed by cylindrical flat-ended AFM tips in lung cells.
Acerbi I, Luque T, Giménez A, Puig M, Reguart N, Farré R, Navajas D, Alcaraz J. Acerbi I, et al. PLoS One. 2012;7(2):e32261. doi: 10.1371/journal.pone.0032261. Epub 2012 Feb 23. PLoS One. 2012. PMID: 22384196 Free PMC article. - Growth factors produced by bone marrow stromal cells on nanoroughened titanium-aluminum-vanadium surfaces program distal MSCs into osteoblasts via BMP2 signaling.
Berger MB, Bosh KB, Jacobs TW, Joshua Cohen D, Schwartz Z, Boyan BD. Berger MB, et al. J Orthop Res. 2021 Sep;39(9):1908-1920. doi: 10.1002/jor.24869. Epub 2020 Oct 12. J Orthop Res. 2021. PMID: 33002223 Free PMC article. - Coordinated Mechanosensitivity of Membrane Rafts and Focal Adhesions.
Fuentes DE, Butler PJ. Fuentes DE, et al. Cell Mol Bioeng. 2012 Jun 1;5(2):143-154. doi: 10.1007/s12195-012-0225-z. Cell Mol Bioeng. 2012. PMID: 23487555 Free PMC article. - Transforming growth factor Beta family: insight into the role of growth factors in regulation of fracture healing biology and potential clinical applications.
Poniatowski ŁA, Wojdasiewicz P, Gasik R, Szukiewicz D. Poniatowski ŁA, et al. Mediators Inflamm. 2015;2015:137823. doi: 10.1155/2015/137823. Epub 2015 Jan 29. Mediators Inflamm. 2015. PMID: 25709154 Free PMC article. Review.
References
- Bazzoni, G., Ma, L., Blue, M. L. and Hemler, M. E. (1998). Divalent cations and ligands induce conformational changes that are highly divergent among β1 integrins. J. Biol. Chem. 273, 6670-6678. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources