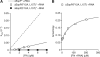Signal sequences activate the catalytic switch of SRP RNA - PubMed (original) (raw)
Signal sequences activate the catalytic switch of SRP RNA
Niels Bradshaw et al. Science. 2009.
Abstract
The signal recognition particle (SRP) recognizes polypeptide chains bearing a signal sequence as they emerge from the ribosome, and then binds its membrane-associated receptor (SR), thereby delivering the ribosome-nascent chain complex to the endoplasmic reticulum in eukaryotic cells and the plasma membrane in prokaryotic cells. SRP RNA catalytically accelerates the interaction of SRP and SR, which stimulates their guanosine triphosphatase (GTPase) activities, leading to dissociation of the complex. We found that although the catalytic activity of SRP RNA appeared to be constitutive, SRP RNA accelerated complex formation only when SRP was bound to a signal sequence. This crucial control step was obscured because a detergent commonly included in the reaction buffer acted as a signal peptide mimic. Thus, SRP RNA is a molecular switch that renders the SRP-SR GTPase engine responsive to signal peptide recruitment, coupling GTP hydrolysis to productive protein targeting.
Figures
Fig. 1
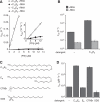
Detergent activates 4.5_S_ RNA to catalyze the Ffh-FtsY interaction. (A) C12E8 stimulates the binding of Ffh and FtsY only in the presence of 4.5_S_ RNA. Observed binding rates for formation of Ffh-FtsY complexes are plotted as a function of Ffh concentration, [Ffh], in the presence and absence of 4.5_S_ RNA and 185 μM C12E8. Lines represent fits to the equation _k_obs = k_off + k_on[Ffh]. Inset shows the slow reactions on an expanded scale. (B) C12E8 activates 4.5_S RNA stimulation of Ffh-FtsY complex dissociation. Dissociation rate constants are plotted in the absence and presence of C12E8. (C) Chemical structures of C12E8, E8, CTABr, and SDS. (D) Association rate constants for Ffh–4.5_S RNA–FtsY complex formation with no detergent, 185 μM C12E8, 100 μM E8, 70 μM CTABr, or 100 μM SDS. Error bars in (B) and (D) are SEs of the fits.
Fig. 2
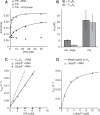
ΔEspP binds SRP with micromolar affinity and stimulates 4.5_S_ RNA catalysis of Ffh-FtsY interaction. (A) Fluorescence anisotropy of ΔEspP-FAM is plotted as a function of [Ffh]. Lines represent fits to the equation Anisotropy = Anisotropyfree + Anisotropybound([Ffh]/(_K_d + [Ffh])). (B) C12E8 increased the K_d of ΔEspP for Ffh–4.5_S RNA. K_d values for ΔEspP binding to Ffh from fluorescence anisotropy in the presence and absence of 4.5_S RNA are plotted. Dark bars represent K_d in the presence of 185 μM C12E8. Error bars are SEs of the fits. (C) In the presence of 4.5_S RNA, ΔEspP stimulates the association rate for Ffh-FtsY complex formation. Observed rate constants are plotted as a function of [Ffh]. Lines are fits to the equation _k_obs = k_off + k_on[Ffh]. The dashed line is a reference to the binding rate in the presence of C12E8 from Fig. 1. (D) ΔEspP* activates 4.5_S RNA by binding to SRP. Observed rates for 1 μM Ffh–4.5_S RNA binding to 1 μM FtsY are plotted as a function of ΔEspP* concentration. The dashed line represents the equation _k_obs = [(fraction bound) (maximum stimulated rate)] + [(fraction unbound)(unstimulated rate)], where the fraction bound was calculated from the _K_d measured in (A); χ2 = 5.4 × 10–6.
Fig. 3
Mutations in ΔEspP that impair SRP-mediated targeting show decreased binding to SRP and decreased stimulation of 4.5_S_ RNA. (A) ΔEspP(F12A, L15T)* stimulates SRP-FtsY complex formation less than does ΔEspP*. The dashed line represents the ΔEspP* + RNA peptide binding rate from Fig. 2C. (B) Fluorescence anisotropy of FAM-labeled ΔEspP bearing Phe12 → Ala and Leu15 → Thr mutations [ΔEspP(F12A, L15T)] is plotted as a function of [Ffh +RNA]. Line is fit as in Fig. 2A.
Similar articles
- Predominant membrane localization is an essential feature of the bacterial signal recognition particle receptor.
Mircheva M, Boy D, Weiche B, Hucke F, Graumann P, Koch HG. Mircheva M, et al. BMC Biol. 2009 Nov 13;7:76. doi: 10.1186/1741-7007-7-76. BMC Biol. 2009. PMID: 19912622 Free PMC article. - SRP RNA controls a conformational switch regulating the SRP-SRP receptor interaction.
Neher SB, Bradshaw N, Floor SN, Gross JD, Walter P. Neher SB, et al. Nat Struct Mol Biol. 2008 Sep;15(9):916-23. doi: 10.1038/nsmb.1467. Nat Struct Mol Biol. 2008. PMID: 19172744 Free PMC article. - RNA-mediated interaction between the peptide-binding and GTPase domains of the signal recognition particle.
Spanggord RJ, Siu F, Ke A, Doudna JA. Spanggord RJ, et al. Nat Struct Mol Biol. 2005 Dec;12(12):1116-22. doi: 10.1038/nsmb1025. Epub 2005 Nov 20. Nat Struct Mol Biol. 2005. PMID: 16299512 - Co-translational protein targeting by the signal recognition particle.
Shan SO, Walter P. Shan SO, et al. FEBS Lett. 2005 Feb 7;579(4):921-6. doi: 10.1016/j.febslet.2004.11.049. FEBS Lett. 2005. PMID: 15680975 Review. - Signal recognition particle: an essential protein-targeting machine.
Akopian D, Shen K, Zhang X, Shan SO. Akopian D, et al. Annu Rev Biochem. 2013;82:693-721. doi: 10.1146/annurev-biochem-072711-164732. Epub 2013 Feb 13. Annu Rev Biochem. 2013. PMID: 23414305 Free PMC article. Review.
Cited by
- Signal sequence-independent SRP-SR complex formation at the membrane suggests an alternative targeting pathway within the SRP cycle.
Braig D, Mircheva M, Sachelaru I, van der Sluis EO, Sturm L, Beckmann R, Koch HG. Braig D, et al. Mol Biol Cell. 2011 Jul 1;22(13):2309-23. doi: 10.1091/mbc.E11-02-0152. Epub 2011 May 5. Mol Biol Cell. 2011. PMID: 21551068 Free PMC article. - Structural insights of non-canonical U*U pair and Hoogsteen interaction probed with Se atom.
Sheng J, Gan J, Soares AS, Salon J, Huang Z. Sheng J, et al. Nucleic Acids Res. 2013 Dec;41(22):10476-87. doi: 10.1093/nar/gkt799. Epub 2013 Sep 5. Nucleic Acids Res. 2013. PMID: 24013566 Free PMC article. - Two-step membrane binding by the bacterial SRP receptor enable efficient and accurate Co-translational protein targeting.
Hwang Fu YH, Huang WYC, Shen K, Groves JT, Miller T, Shan SO. Hwang Fu YH, et al. Elife. 2017 Jul 28;6:e25885. doi: 10.7554/eLife.25885. Elife. 2017. PMID: 28753124 Free PMC article. - Fingerloop activates cargo delivery and unloading during cotranslational protein targeting.
Ariosa AR, Duncan SS, Saraogi I, Lu X, Brown A, Phillips GJ, Shan SO. Ariosa AR, et al. Mol Biol Cell. 2013 Jan;24(2):63-73. doi: 10.1091/mbc.E12-06-0434. Epub 2012 Nov 7. Mol Biol Cell. 2013. PMID: 23135999 Free PMC article. - Membrane proteins take center stage in Frankfurt.
Schleiff E, Tampé R. Schleiff E, et al. Nat Chem Biol. 2009 Mar;5(3):135-9. doi: 10.1038/nchembio0309-135. Nat Chem Biol. 2009. PMID: 19219012
References
- Keenan RJ, Freymann DM, Stroud RM, Walter P. Annu. Rev. Biochem. 2001;70:755. - PubMed
- Miller JD, Wilhelm H, Gierasch L, Gilmore R, Walter P. Nature. 1993;366:351. - PubMed
- Peluso P, Shan SO, Nock S, Herschlag D, Walter P. Biochemistry. 2001;40:15224. - PubMed
- Powers T, Walter P. Science. 1995;269:1422. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM032384/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 GM32384/GM/NIGMS NIH HHS/United States
- R25 GM56847/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials