Viral sensors: diversity in pathogen recognition - PubMed (original) (raw)
Review
Viral sensors: diversity in pathogen recognition
Stephen A McCartney et al. Immunol Rev. 2009 Jan.
Abstract
Innate sensors of viral infection detect viral products and initiate the signal cascades that lead to the antiviral response. Several proteins have been identified to play a role in this process, mostly members of the Toll-like receptor and retinoic acid-inducible gene I-like receptor families. These receptors have been demonstrated to function in part by recognizing a diverse yet unique repertoire of nucleic acid substrates. Upon recognition of their ligands, these sensors activate distinct signaling pathways that lead to the secretion of type I interferon and inflammatory cytokines. It remains to be seen, however, if these sensors are redundant or whether each serves a unique function. In this work, we review the current knowledge of viral sensors, speculate on how they may function in vivo, and explore the potential reasons for their diversity.
Figures
Figure 1
Cytoplasmic and endosomal sensors of viral nucleic acids. This figure illustrates the detection of viral products by retinoic acid‐inducible gene I (RIG‐I)‐like receptor (RLR) and Toll‐like receptor (TLR) family members. TLR3, TLR7/ 8, and TLR9 are located on endosomal compartments in which they sense their double‐stranded RNA (dsRNA), single‐stranded RNA (ssRNA), and CpG DNA ligands, respectively. TLR3 signals through the adapter protein Toll/interleukin‐1 (IL‐1) receptor (TIR) domain‐containing adapter‐inducing interferon‐β (TRIF), which activates tumor necrosis factor (TNF) receptor‐associated factor 3 (TRAF3) (not shown) and the TANK‐binding kinase 1 (TBK‐1) complex leading to interferon (IFN) regulatory factor 7 (IRF‐7) activation and IFN signaling. TRIF also signals through TRAF6, which leads to nuclear factor κB (NF‐κB) activation and inflammatory cytokine production. TLR7, TLR8, and TLR9 signal through the MyD88 adapter. MyD88 signals through a protein complex consisting of TRAF6 and IL‐1 receptor‐associated kinase 1/4 (IRAK1/4) (not shown), leading to the activation of type I IFN and NF‐κB signaling. RLR family members, melanoma differentiation‐associated gene 5 (MDA5), retinoic acid‐inducible gene I (RIG‐I), and laboratory of genetics and physiology‐2 (LGP2), are cytoplasmic proteins that detect viral products within the cytosol. MDA5 and RIG‐I signal through IFN‐β promoter stimulator 1 (IPS‐1), which is located on the mitochondrial membrane. IPS‐1 signals through TRAF3 and the TBK‐1/inhibitor of NF‐κB kinase ε complex to activate IRF‐3 and IRF‐7 and then type I IFN. IPS‐1 also signals through FAS‐associated death domain‐containing protein (FADD) leading to the activation of caspase‐8 and caspase‐10 (not shown), which causes NF‐κB activation and inflammatory cytokine production. LGP2 does not signal through IPS‐1 and is considered to be a negative regulator of RIG‐I.
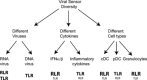
Figure 2
Hypothetical model of functional diversity among viral sensors. Three potential mechanisms by which viral sensors perform unique functions: (i) viral sensors detect different ligands resulting in the recognition of a variety of viral families and/or the recognition of different components of the same virus. (ii) Toll‐like receptor (TLR) and retinoic acid‐inducible gene I‐like receptor (RLR) signaling results in the production of different cytokine responses (i.e. type I interferon versus pro‐inflammatory cytokines). (iii) The expression of TLRs and RLRs in different cell types results in unique responses to viruses in different cells and tissues. Predominant sensors are indicated in bold, while secondary sensors are indicated below in smaller font.
Similar articles
- RNA recognition and signal transduction by RIG-I-like receptors.
Yoneyama M, Fujita T. Yoneyama M, et al. Immunol Rev. 2009 Jan;227(1):54-65. doi: 10.1111/j.1600-065X.2008.00727.x. Immunol Rev. 2009. PMID: 19120475 Review. - Recognition of viral nucleic acids in innate immunity.
Yoneyama M, Fujita T. Yoneyama M, et al. Rev Med Virol. 2010 Jan;20(1):4-22. doi: 10.1002/rmv.633. Rev Med Virol. 2010. PMID: 20041442 Review. - How Dengue Virus Circumvents Innate Immunity.
Kao YT, Lai MMC, Yu CY. Kao YT, et al. Front Immunol. 2018 Dec 4;9:2860. doi: 10.3389/fimmu.2018.02860. eCollection 2018. Front Immunol. 2018. PMID: 30564245 Free PMC article. Review. - Identifying RNA Sensors in Antiviral Innate Immunity.
Zheng C, Zhang L. Zheng C, et al. Methods Mol Biol. 2025;2854:107-115. doi: 10.1007/978-1-0716-4108-8_12. Methods Mol Biol. 2025. PMID: 39192123 - Functional evolution of the TICAM-1 pathway for extrinsic RNA sensing.
Seya T, Matsumoto M, Ebihara T, Oshiumi H. Seya T, et al. Immunol Rev. 2009 Jan;227(1):44-53. doi: 10.1111/j.1600-065X.2008.00723.x. Immunol Rev. 2009. PMID: 19120474 Review.
Cited by
- A Guide to Nucleic Acid Vaccines in the Prevention and Treatment of Infectious Diseases and Cancers: From Basic Principles to Current Applications.
Qin F, Xia F, Chen H, Cui B, Feng Y, Zhang P, Chen J, Luo M. Qin F, et al. Front Cell Dev Biol. 2021 May 25;9:633776. doi: 10.3389/fcell.2021.633776. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34113610 Free PMC article. Review. - Type I IFN-mediated regulation of IL-1 production in inflammatory disorders.
Ludigs K, Parfenov V, Du Pasquier RA, Guarda G. Ludigs K, et al. Cell Mol Life Sci. 2012 Oct;69(20):3395-418. doi: 10.1007/s00018-012-0989-2. Epub 2012 Apr 24. Cell Mol Life Sci. 2012. PMID: 22527721 Free PMC article. Review. - TLR3 activation efficiency by high or low molecular mass poly I:C.
Zhou Y, Guo M, Wang X, Li J, Wang Y, Ye L, Dai M, Zhou L, Persidsky Y, Ho W. Zhou Y, et al. Innate Immun. 2013;19(2):184-92. doi: 10.1177/1753425912459975. Epub 2012 Oct 3. Innate Immun. 2013. PMID: 23035017 Free PMC article. - Bacteriophage and the Innate Immune System: Access and Signaling.
Carroll-Portillo A, Lin HC. Carroll-Portillo A, et al. Microorganisms. 2019 Nov 28;7(12):625. doi: 10.3390/microorganisms7120625. Microorganisms. 2019. PMID: 31795262 Free PMC article. Review. - Mechanism of inhibiting type I interferon induction by hepatitis B virus X protein.
Jiang J, Tang H. Jiang J, et al. Protein Cell. 2010 Dec;1(12):1106-17. doi: 10.1007/s13238-010-0141-8. Epub 2011 Jan 8. Protein Cell. 2010. PMID: 21213104 Free PMC article.
References
- Takeuchi O, Akira S. Recognition of viruses by innate immunity. Immunol Rev 2007;220:214–224. - PubMed
- Yoneyama M, et al. The RNA helicase RIG‐I has an essential function in double‐stranded RNA‐induced innate antiviral responses. Nat Immunol 2004;5:730–737. - PubMed
- Kovacsovics M, et al. Overexpression of Helicard, a CARD‐containing helicase cleaved during apoptosis, accelerates DNA degradation. Curr Biol 2002;12:838–843. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources