Microbe sensing, positive feedback loops, and the pathogenesis of inflammatory diseases - PubMed (original) (raw)
Review
Microbe sensing, positive feedback loops, and the pathogenesis of inflammatory diseases
Bruce Beutler. Immunol Rev. 2009 Jan.
Abstract
The molecular apparatus that protects us against infection can also injure us by causing autoimmune or autoinflammatory disease. It now seems that at times, defects within the sensing arm of innate immunity contribute to diseases of this type. The initiation of an immune response is often microbe dependent and, in many cases, Toll-like receptor (TLR) dependent. Positive feedback loops triggering immune activation may occur when TLR signaling pathways stimulate host cells in an unchecked manner. Or, immune activation may persist because of failure to eradicate an inciting infection. Or on occasion, endogenous DNA may trigger specific immune responses that beget further responses in a TLR-dependent autoamplification loop. Specific biochemical defects that cause loop-related autoimmunity have been revealed by random germline mutagenesis and by gene targeting. We have also developed some insight into critical points at which feedback loops can be interrupted.
Figures
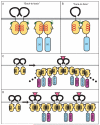
Fig. 1. TLR:MyD88 interactions
The interfaces between two receptors or two adapters or receptor and adapter are predicted from docking studies based on the structures of the TIR domains of TLR1 and TLR2. (A) Binding between two receptors or two adapters may be mediated by ‘back-to-back’ TIR domain (light orange) interactions, which are characterized by homotypic interactions between αE helices (red). The back-to-back interface does not mediate receptor:adapter binding. (B) ‘Face-to-face’ TIR domain interactions likely mediate receptor:adapter binding and involve the BB loop (blue) and Poc site (green) from each molecule. Whether face-to-face interactions can also mediate receptor:receptor or adapter:adapter binding is untested but expected based on molecular docking studies. (C and D) Models for recruitment and ‘chaining’ of adapter molecules upon TLR activation. In (C), ligand binding causes dimerization of two TLRs in a back-to-back fashion, followed by recruitment of adapter (MyD88) to the receptor by a face-to-face interaction. Signal amplification may be achieved by the subsequent recruitment of additional adapters (MyD88 and MAL), which bind each other with alternating face-to-face and back-to-back interactions. Alternatively, as shown in (D), upon ligand binding and back-to-back dimerization of receptors, each receptor complex recruits one adapter by a face-to-face interaction. Adjacent adapters (MyD88 and MAL) then form back-to-back interfaces.
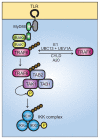
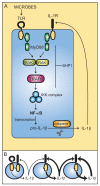
Fig. 2. Ubiquitin signaling in TLR-mediated activation of NF-κB
Ligand binding to TLRs leads to the recruitment of MyD88 and, subsequently, IRAK4 and IRAK1. IRAK1 binds to TRAF6. The oligomerization of TRAF6 (not shown) activates its ubiquitin ligase activity, leading to K63-linked polyubiquitination of NEMO and TRAF6 in conjunction with UBC13 and UEV1A. De-ubiquitination can be carried out by CYLD and A20. Ubiquitinated TRAF6 recruits the TRIKA2 complex, consisting of TAB1, TAB2, and TAK1, via binding to the novel zinc finger domain of TAB2. TAK1 kinase becomes activated by autophosphorylation and phosphorylates IKKβ at two activation loop serine residues, resulting in IKK complex activation. The IKK complex then phosphorylates IκB, leading to IκB degradation, and releasing NF-κB for translocation to the nucleus.
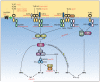
Fig. 3. Overview of the TLR signaling pathways
The pathways represent data obtained from both forward and reverse genetic studies. ENU-induced phenotypes are shown in red text. Note that TLR3, TLR7, and T:R9 are endosomal proteins, while TLR1, TLR2, TLR6, TLR4, as well as TLR5 (not shown) are expressed on the plasma membrane. Unc93b1 is an endoplasmic reticulum protein that influences endosome function. TLR activation recruits TRIF, TRAM, MAL, and/or MyD88 and leads to the activation of IRAK1 and IRAK4, or the TBK1 or IKKι family kinases. TRAF6 signaling to the TAB1/TAB2/TAK1 complex results in the activation of NF-κB and AP-1, via the functions of the IKK complex, Tpl2, and JNK. IRF5 is another means by which signaling through MyD88 leads to TNF production. MyD88 and TRAF6 may interact directly with IRF5 in a complex, activating IRF5 and promoting its translocation to the nucleus. Additionally, MyD88, together with TRAF6 and IRAK4, has also been shown to bind IRF7 directly in order to stimulate IFN-α production. This occurs downstream of TLR7, TLR8 and TLR9 in plasmacytoid dendritic cells (pDCs) and requires the phosphorylation of IRF7 by IRAK1. TRAF3 is also involved in TLR7- and TLR9-dependent pDC production of IFN-α; TRAF3 may form a complex with IRAK1 and IRF7 and facilitate phosphorylation of IRF7 (42) (not shown). In the MyD88-independent pathway, TLR3 or TLR4 recruit TRIF and TBK1, the critical kinase required for activation of IRF3. TRAF3 mediates the interaction between TRIF and TBK1. The kinase RIP-1 is required for NF-κB activation downstream of TRIF, and likely requires the direct interaction between TRIF and RIP-1. The point at which RIP-1 signaling impinges on the NF-κB pathway is unknown (dashed gray arrow).
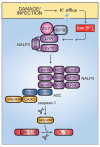
Fig. 4. IL-1β activation by the NALP3 inflammasome
Activation of the NALP3 inflammasome is triggered by a wide variety of stimuli, all of which may be united in their ability to cause K+ efflux from the host cell. Low intracellular K+ concentrations lead, through an unknown mechanism, to dissociation of HSP90 and SGT1 from the autorepressed NALP3. NALP3 can then recruit ASC via pyrin-pyrin domain interactions. ASC in turn recruits caspase 1 via CARD-CARD domain binding. Together, NALP3, ASC, and caspase 1 form the core components of the NALP3 inflammasome. Processing of caspase 1 activates it to cleave pro-IL-1β, giving rise to active IL-1β.
Fig. 5. RLH signaling
The two cytoplasmic nucleic acid sensors, RIG-I and MDA5, respond to different viral infections, recognizing 5′-triphosphate single stranded RNA and dsRNA, respectively. RIG-I also detects dsRNA. RIG-I and MDA5 activate IPS1, which recruits TRAF3 and FADD into a complex. Association of FADD with pro-caspase 8 or pro-caspase 10 results in cleavage to the mature, active caspase 8 or caspase 10, which go on to activate NF-κB. RIG-I and MDA5 signaling also activates IRF3 and IRF7. This occurs through the kinases IKKι and TBK. K63-linked ubiquitination of RIG-I by TRIM25 enhances RIG-I signaling.
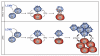
Fig. 6. HLH-like disease in Unc13djinx mice is caused by an uninhibited cytokine production loop
In WT animals, LCMV-infected antigen-presenting cells (APCs) present antigen to CD8+ T cells, stimulating them to produce IFNγ, which may further promote antigen presentation. Degranulation by CD8+ T cells halts this cycle by killing the target APCs. In Unc13djinx mice, however, lymphoid cells fail to degranulate and kill LCMV-infected targets, which continue to shed virus and infect other cells. Persistent production of IFNγ drives the expansion of myeloid cells (not shown), which present antigen and further stimulate lymphoid cell proliferation and activation. The viral pathogen is never cleared, and mice develop clinical features of HLH, including thrombocytopenia, neutrophilia, splenomegaly, and elevated serum IFN-γ.
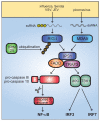
Fig. 7. Unchecked IL-1 signaling causes inflammation and autoimmunity in Ptpn6spin mice
(A) The IL-1 signaling loop is initiated by microbial stimulation of TLRs, which through canonical TLR signaling via IRAK1, IRAK4, TRAF6, IKK complex, and NF-κB activation, leads to IL-1β production. IL-1β activates its own receptor, which also signals via the same TLR pathway components to produce IL-1β. Through an unknown mechanism, impaired SHP1 function in Ptpn6spin cells results in a failure to downregulate IL-1 signaling, which continues to stimulate its own production. This uninhibited IL-1 signaling loop (B) ultimately causes chronic inflammation and autoimmunity in Ptpn6spin mice. Derivation into a germfree environment and inactivating mutations in MyD88, IRAK4, and IL-1R suppress disease in Ptpn6spin mutants.
Similar articles
- Activation of Both TLR and NOD Signaling Confers Host Innate Immunity-Mediated Protection Against Microbial Infection.
Zhou H, Coveney AP, Wu M, Huang J, Blankson S, Zhao H, O'Leary DP, Bai Z, Li Y, Redmond HP, Wang JH, Wang J. Zhou H, et al. Front Immunol. 2019 Jan 14;9:3082. doi: 10.3389/fimmu.2018.03082. eCollection 2018. Front Immunol. 2019. PMID: 30692992 Free PMC article. - Synergistic Activation of Toll-Like and NOD Receptors by Complementary Antigens as Facilitators of Autoimmune Disease: Review, Model and Novel Predictions.
Root-Bernstein R. Root-Bernstein R. Int J Mol Sci. 2020 Jun 30;21(13):4645. doi: 10.3390/ijms21134645. Int J Mol Sci. 2020. PMID: 32629865 Free PMC article. Review. - Host-pathogen interactions.
Kaisho T, Wagner H. Kaisho T, et al. Curr Opin Immunol. 2008 Aug;20(4):369-70. doi: 10.1016/j.coi.2008.07.003. Epub 2008 Jul 30. Curr Opin Immunol. 2008. PMID: 18625309 No abstract available. - Toll-like receptors (TLRs) and Nod-like receptors (NLRs) in inflammatory disorders.
Fukata M, Vamadevan AS, Abreu MT. Fukata M, et al. Semin Immunol. 2009 Aug;21(4):242-53. doi: 10.1016/j.smim.2009.06.005. Semin Immunol. 2009. PMID: 19748439 Review. - Toll-like receptors and NOD-like receptors: domain architecture and cellular signalling.
Langefeld T, Mohamed W, Ghai R, Chakraborty T. Langefeld T, et al. Adv Exp Med Biol. 2009;653:48-57. doi: 10.1007/978-1-4419-0901-5_4. Adv Exp Med Biol. 2009. PMID: 19799111 Review.
Cited by
- The Effect of Orally Administered Multi-Strain Probiotic Formulation (Lactobacillus, Bifidobacterium) on the Phagocytic Activity and Oxidative Metabolism of Peripheral Blood Granulocytes and Monocytes in Lambs.
Wójcik R, Małaczewska J, Tobolski D, Miciński J, Kaczorek-Łukowska E, Zwierzchowski G. Wójcik R, et al. Int J Mol Sci. 2024 May 7;25(10):5068. doi: 10.3390/ijms25105068. Int J Mol Sci. 2024. PMID: 38791112 Free PMC article. - Side effects of CoronaVac® COVID-19 vaccination: Investigation in North Jakarta district public health center communities in Indonesia.
Ramatillah DL, Gan SH, Novarticia J, Araminda GN, Michael M, Elnaem M, Alawuddin R, Khan K. Ramatillah DL, et al. Heliyon. 2024 Apr 26;10(9):e30087. doi: 10.1016/j.heliyon.2024.e30087. eCollection 2024 May 15. Heliyon. 2024. PMID: 38694099 Free PMC article. - Shedding light on the molecular and regulatory mechanisms of TLR4 signaling in endothelial cells under physiological and inflamed conditions.
Stierschneider A, Wiesner C. Stierschneider A, et al. Front Immunol. 2023 Nov 24;14:1264889. doi: 10.3389/fimmu.2023.1264889. eCollection 2023. Front Immunol. 2023. PMID: 38077393 Free PMC article. Review. - Longitudinal changes in tear cytokines and antimicrobial proteins in trachomatous disease.
Barton A, Faal N, Ramadhani A, Derrick T, Mafuru E, Mtuy T, Massae P, Malissa A, Joof H, Makalo P, Sillah A, Harte A, Pickering H, Bailey R, Mabey DC, Burton MJ, Holland MJ. Barton A, et al. PLoS Negl Trop Dis. 2023 Oct 20;17(10):e0011689. doi: 10.1371/journal.pntd.0011689. eCollection 2023 Oct. PLoS Negl Trop Dis. 2023. PMID: 37862368 Free PMC article. - Covid 19 vaccines and the misinterpretation of perceived side effects clarity on the safety of vaccines.
Palmer RD. Palmer RD. Biomedicine (Taipei). 2022 Sep 1;12(3):1-4. doi: 10.37796/2211-8039.1371. eCollection 2022. Biomedicine (Taipei). 2022. PMID: 36381188 Free PMC article.
References
- Runza VL, Schwaeble W, Mannel DN. Ficolins: novel pattern recognition molecules of the innate immune response. Immunobiology. 2008;213:297–306. - PubMed
- Schwaeble WJ, Reid KB. Does properdin crosslink the cellular and the humoral immune response? Immunol Today. 1999;20:17–21. - PubMed
- Boswell RN, Austen KF, Goetzl EJ. A chemotactic receptor for val(ala)-gly-ser-glu on human. Immunol Commun. 1976;5:469–479. - PubMed
- Sultzer BM. Genetic control of leucocyte responses to endotoxin. Nature. 1968;219:1253–1254. - PubMed
- Beutler B. Neo-ligands for innate immune receptors and the etiology of sterile inflammatory disease. Immunol Rev. 2007;220:113–128. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical