Upstream sequence elements direct post-transcriptional regulation of gene expression under stress conditions in yeast - PubMed (original) (raw)
Upstream sequence elements direct post-transcriptional regulation of gene expression under stress conditions in yeast
Craig Lawless et al. BMC Genomics. 2009.
Abstract
Background: The control of gene expression in eukaryotic cells occurs both transcriptionally and post-transcriptionally. Although many genes are now known to be regulated at the translational level, in general, the mechanisms are poorly understood. We have previously presented polysomal gradient and array-based evidence that translational control is widespread in a significant number of genes when yeast cells are exposed to a range of stresses. Here we have re-examined these gene sets, considering the role of UTR sequences in the translational responses of these genes using recent large-scale datasets which define 5' and 3' transcriptional ends for many yeast genes. In particular, we highlight the potential role of 5' UTRs and upstream open reading frames (uORFs).
Results: We show a highly significant enrichment in specific GO functional classes for genes that are translationally up- and down-regulated under given stresses (e.g. carbohydrate metabolism is up-regulated under amino acid starvation). Cross-referencing these data with the stress response data we show that translationally upregulated genes have longer 5' UTRs, consistent with their role in translational regulation. In the first genome-wide study of uORFs in a set of mapped 5' UTRs, we show that uORFs are rare, being statistically under-represented in UTR sequences. However, they have distinct compositional biases consistent with their putative role in translational control and are more common in genes which are apparently translationally up-regulated.
Conclusion: These results demonstrate a central regulatory role for UTR sequences, and 5' UTRs in particular, highlighting the significant role of uORFs in post-transcriptional control in yeast. Yeast uORFs are more highly conserved than has been suggested, lending further weight to their significance as functional elements involved in gene regulation. It also suggests a more complex and novel mechanism of control, whereby uORFs permit genes to escape from a more general attenuation of translation under conditions of stress. However, since uORFs are relatively rare (only ~13% of yeast genes have them) there remain many unanswered questions as to how UTR elements can direct translational control of many hundreds of genes under stress.
Figures
Figure 1
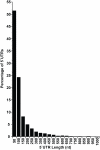
Length distribution of yeast 5' UTRs. Data was taken from a superset of transcriptional start sites, selecting one per gene. The majority are less than 50 nucleotides in length.
Figure 2
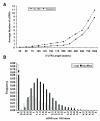
Statistical significance in yeast upstream sequences and 5' UTRs. In A), the average number of uORFs is shown for both upstream regions in general (No TSS) and TSS mapped 5' UTRs (Superset). The same number of sequences (4149) is used in all cases for the "No TSS" line. At all points, there are fewer uORFs in the real 5' UTR sequences, In B), a normalised frequency histogram of uORF numbers in real and shuffled 5' UTRs highlights the reduced chance of observing a uORF in real UTR sequence compared to random. Over 85% have none compared to 68% by chance.
Figure 3
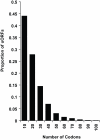
Size of uORFs in 5' UTRs. The distribution of uORF lengths in the mapped 5' UTRs is shown; the vast majority are shorter than 20 codons.
Figure 4
Relative uORF compositional adaption to the translational machinery. Panel A shows the frequency histogram of log2 ratios of uORF to principal ORF start codon adaption scores (AUG-CAI(r)). If a uORF is better adapted than the principal ORF, it will produce a positive log value. Panel B shows that same comparison considering the translation adpation index (tAI) of the complete reading frame in order to calculate the log2 ratio. In both cases, uORFs are considerably less well adapted in general with log2 mean values less than zero.
Figure 5
GoStat over-representation statistics for translationally controlled gene subsets under amino acid starvation. The numbers of genes in each GO functional category are shown for yeast genes which are differentially translationally regulated under amino acid starvation conditions. Panel A shows significant over-representation for genes which are up-regulated, and panel B for those that are down-regulated. Panel C shows the greyscale key for the associated p-values. Only GO categories with p-values < 0.001 are shown.
Figure 6
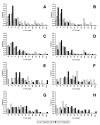
Histograms of UTR lengths for translationally controlled gene sets. The 5' UTR lengths are shown for gene sets that are up- or down- regulated under varying stress conditions. A. Amino acid starvation, B. Butanol addition, C. 0.2 mM H2O2, D, 2 mM H2O2., along with 3' UTR length distributions for gene sets under E. Amino acid starvation, F. Butanol addition, G. 0.2 mM H2O2, and H, 2 mM H2O2
Figure 7
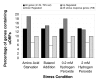
Proportion of uORF-containing genes in translationally controlled gene sets. The fraction of different gene sets whose UTRs contain uORFs are shown for different stress conditions labelled on the x-axis. This is compared to all 4149 TSS-mapped genes and all 708 genes showing differential regulation in any of the 4 stress conditions. A general trend of uORF enrichment is observed in the up-regulated gene sets compared to down-regulated under all stresses. Up and down-regulated genes were excluded from the "All Genes" set.
Figure 8
Conservation statistics for highly conserved uORFs. For individual uORFs with mean phastCons scores greater than 0.99 (highly conserved regions) across 6 Sensu Stricto yeast species, the phastCons score is plotted against corresponding number of species (out of 6) in which the uORF is conserved. This is shown with respect to an "average" species conservation metric comparing sequences with similar properties to an open reading frame (see Methods). These 61 uORFs show considerable conservation, both in terms of local sequence context (phastCons scores) and the number of species which also share a uORF, and includes previously characterised uORFs from the well known GCN4 gene, shown as circled dots.
Similar articles
- Identification and characterization of upstream open reading frames (uORF) in the 5' untranslated regions (UTR) of genes in Saccharomyces cerevisiae.
Zhang Z, Dietrich FS. Zhang Z, et al. Curr Genet. 2005 Aug;48(2):77-87. doi: 10.1007/s00294-005-0001-x. Epub 2005 Sep 14. Curr Genet. 2005. PMID: 16012843 - Extensive transcript diversity and novel upstream open reading frame regulation in yeast.
Waern K, Snyder M. Waern K, et al. G3 (Bethesda). 2013 Feb;3(2):343-52. doi: 10.1534/g3.112.003640. Epub 2013 Feb 1. G3 (Bethesda). 2013. PMID: 23390610 Free PMC article. - Dual modes of natural selection on upstream open reading frames.
Neafsey DE, Galagan JE. Neafsey DE, et al. Mol Biol Evol. 2007 Aug;24(8):1744-51. doi: 10.1093/molbev/msm093. Epub 2007 May 9. Mol Biol Evol. 2007. PMID: 17494029 - A helicase links upstream ORFs and RNA structure.
Jankowsky E, Guenther UP. Jankowsky E, et al. Curr Genet. 2019 Apr;65(2):453-456. doi: 10.1007/s00294-018-0911-z. Epub 2018 Nov 27. Curr Genet. 2019. PMID: 30483885 Free PMC article. Review. - Regulation of plant translation by upstream open reading frames.
von Arnim AG, Jia Q, Vaughn JN. von Arnim AG, et al. Plant Sci. 2014 Jan;214:1-12. doi: 10.1016/j.plantsci.2013.09.006. Epub 2013 Sep 18. Plant Sci. 2014. PMID: 24268158 Review.
Cited by
- An MSC2 Promoter-lacZ Fusion Gene Reveals Zinc-Responsive Changes in Sites of Transcription Initiation That Occur across the Yeast Genome.
Wu YH, Taggart J, Song PX, MacDiarmid C, Eide DJ. Wu YH, et al. PLoS One. 2016 Sep 22;11(9):e0163256. doi: 10.1371/journal.pone.0163256. eCollection 2016. PLoS One. 2016. PMID: 27657924 Free PMC article. - Transcriptional responses of Saccharomyces cerevisiae to shift from respiratory and respirofermentative to fully fermentative metabolism.
Rintala E, Jouhten P, Toivari M, Wiebe MG, Maaheimo H, Penttilä M, Ruohonen L. Rintala E, et al. OMICS. 2011 Jul-Aug;15(7-8):461-76. doi: 10.1089/omi.2010.0082. Epub 2011 Feb 24. OMICS. 2011. PMID: 21348598 Free PMC article. - Upstream ORFs are prevalent translational repressors in vertebrates.
Johnstone TG, Bazzini AA, Giraldez AJ. Johnstone TG, et al. EMBO J. 2016 Apr 1;35(7):706-23. doi: 10.15252/embj.201592759. Epub 2016 Feb 19. EMBO J. 2016. PMID: 26896445 Free PMC article. - eIF4E-binding protein regulation of mRNAs with differential 5'-UTR secondary structure: a polyelectrostatic model for a component of protein-mRNA interactions.
Cawley A, Warwicker J. Cawley A, et al. Nucleic Acids Res. 2012 Sep;40(16):7666-75. doi: 10.1093/nar/gks511. Epub 2012 Jun 20. Nucleic Acids Res. 2012. PMID: 22718971 Free PMC article. - Before It Gets Started: Regulating Translation at the 5' UTR.
Araujo PR, Yoon K, Ko D, Smith AD, Qiao M, Suresh U, Burns SC, Penalva LO. Araujo PR, et al. Comp Funct Genomics. 2012;2012:475731. doi: 10.1155/2012/475731. Epub 2012 May 28. Comp Funct Genomics. 2012. PMID: 22693426 Free PMC article.
References
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KT, Lander ES, Gifford DK, Fraenkel E, Young RA. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/D006996/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBSSA200410904/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases