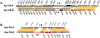Germ warfare in a microbial mat community: CRISPRs provide insights into the co-evolution of host and viral genomes - PubMed (original) (raw)
Germ warfare in a microbial mat community: CRISPRs provide insights into the co-evolution of host and viral genomes
John F Heidelberg et al. PLoS One. 2009.
Abstract
CRISPR arrays and associated cas genes are widespread in bacteria and archaea and confer acquired resistance to viruses. To examine viral immunity in the context of naturally evolving microbial populations we analyzed genomic data from two thermophilic Synechococcus isolates (Syn OS-A and Syn OS-B') as well as a prokaryotic metagenome and viral metagenome derived from microbial mats in hotsprings at Yellowstone National Park. Two distinct CRISPR types, distinguished by the repeat sequence, are found in both the Syn OS-A and Syn OS-B' genomes. The genome of Syn OS-A contains a third CRISPR type with a distinct repeat sequence, which is not found in Syn OS-B', but appears to be shared with other microorganisms that inhabit the mat. The CRISPR repeats identified in the microbial metagenome are highly conserved, while the spacer sequences (hereafter referred to as "viritopes" to emphasize their critical role in viral immunity) were mostly unique and had no high identity matches when searched against GenBank. Searching the viritopes against the viral metagenome, however, yielded several matches with high similarity some of which were within a gene identified as a likely viral lysozyme/lysin protein. Analysis of viral metagenome sequences corresponding to this lysozyme/lysin protein revealed several mutations all of which translate into silent or conservative mutations which are unlikely to affect protein function, but may help the virus evade the host CRISPR resistance mechanism. These results demonstrate the varied challenges presented by a natural virus population, and support the notion that the CRISPR/viritope system must be able to adapt quickly to provide host immunity. The ability of metagenomics to track population-level variation in viritope sequences allows for a culture-independent method for evaluating the fast co-evolution of host and viral genomes and its consequence on the structuring of complex microbial communities.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
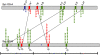
Figure 1. Schematic location of CRISPR loci on Syn OS-A OS-B genomes.
Bars indicate the relative positions of the Type I (red), Type II (green) and Type III (blue) CRISPR loci on the genomes of Syn OS-A (top) and Syn OS-B′ (bottom). Asterisks indicate CRISPRs with an associated cas operon. CRISPRs within syntenic genome blocks are connected with lines. Approximate location and direction of the mapped clone-mates of CRISPR containing clones are show as triangles colored by the repeat type (as above). Two clones that mapped to a location on a reference genome that lacks a CRISPR array are shown within the genome box. Line below shows genome size (large ticks at 1 Mbp; small ticks at 0.5 Mbp).
Figure 2. Type I CRISPR regions.
A) CRISPR-IA region from Syn OS-A (top) and Syn OS-B′ (bottom). The cas gene cluster is indicated by a grey bracket. B) CRISPR-IB region. Gene identifiers are shown above or below each gene, excluding the GenBank locus tag prefix ‘CYA_’ (for Syn OS-A) or ‘CYB_’ (for Syn OS-B′). Genes are color-coded following the COG-based IMG convention (
), (described in Figure 4), except that genes annotated as “hypothetical protein” are in grey, genes with no putative ortholog in the other genome are in red, and genes annotated as transposases are in black. Orthologous genes are indicated by yellow blocks. Detailed information on each gene is available in Table S3.
Figure 3. Type II CRISPR regions.
Homologous regions from Syn OS-A and Syn OS-B′ where one or both genomes has a Type II CRISPR locus are displayed. The CRISPR displayed is indicated by the panel letter (e.g. panel ‘A’ shows CRISPR-IIA, panel ‘B’ shows CRISPR-IIB, etc.) Figure conventions are as described in Figure 2. FS indicates a frameshift in the CDS. Additional details can be found in Table S3.
Figure 4. Comparison of Type III CRISPR locus to Roseiflexus RS-1 and Symbiobacterium.
Homologous regions from Syn OS-A, _Syn_-OS-B′, Roseiflexus RS-1 and Symbiobacterium are displayed. Gene identifiers are shown above or below each gene, excluding the GenBank locus tag prefix ‘CYA_’ (for Syn OS-A), ‘CYB_’ (for Syn OS-B′), RoseRS_ (for Roseiflexus RS-1) or STH (for Symbiobacterium thermophilum). Additional figure conventions are as described in Figure 2. Details about the genes can be found in Table S3.
Figure 5. Example of a putative gene encoding a lysozyme derived from the virome.
The location of the CRISPR viritopes is highlighted. The yellow highlighted region matches CRISPR_II_YMBCR81TF-SP-2 and the bold highlighted region is CRISPR_II_YMIA938TF-SP-4/5.
Similar articles
- Diverse CRISPRs evolving in human microbiomes.
Rho M, Wu YW, Tang H, Doak TG, Ye Y. Rho M, et al. PLoS Genet. 2012;8(6):e1002441. doi: 10.1371/journal.pgen.1002441. Epub 2012 Jun 13. PLoS Genet. 2012. PMID: 22719260 Free PMC article. - The CRISPR Spacer Space Is Dominated by Sequences from Species-Specific Mobilomes.
Shmakov SA, Sitnik V, Makarova KS, Wolf YI, Severinov KV, Koonin EV. Shmakov SA, et al. mBio. 2017 Sep 19;8(5):e01397-17. doi: 10.1128/mBio.01397-17. mBio. 2017. PMID: 28928211 Free PMC article. - Major bacterial lineages are essentially devoid of CRISPR-Cas viral defence systems.
Burstein D, Sun CL, Brown CT, Sharon I, Anantharaman K, Probst AJ, Thomas BC, Banfield JF. Burstein D, et al. Nat Commun. 2016 Feb 3;7:10613. doi: 10.1038/ncomms10613. Nat Commun. 2016. PMID: 26837824 Free PMC article. - CRISPR/Cas, the immune system of bacteria and archaea.
Horvath P, Barrangou R. Horvath P, et al. Science. 2010 Jan 8;327(5962):167-70. doi: 10.1126/science.1179555. Science. 2010. PMID: 20056882 Review. - Flexible genomic islands as drivers of genome evolution.
Rodriguez-Valera F, Martin-Cuadrado AB, López-Pérez M. Rodriguez-Valera F, et al. Curr Opin Microbiol. 2016 Jun;31:154-160. doi: 10.1016/j.mib.2016.03.014. Epub 2016 Apr 14. Curr Opin Microbiol. 2016. PMID: 27085300 Review.
Cited by
- Dynamics in microbial communities: unraveling mechanisms to identify principles.
Konopka A, Lindemann S, Fredrickson J. Konopka A, et al. ISME J. 2015 Jul;9(7):1488-95. doi: 10.1038/ismej.2014.251. Epub 2014 Dec 19. ISME J. 2015. PMID: 25526370 Free PMC article. - Modeling microbial community structure and functional diversity across time and space.
Larsen PE, Gibbons SM, Gilbert JA. Larsen PE, et al. FEMS Microbiol Lett. 2012 Jul;332(2):91-8. doi: 10.1111/j.1574-6968.2012.02588.x. Epub 2012 May 28. FEMS Microbiol Lett. 2012. PMID: 22553907 Free PMC article. Review. - Hot spring metagenomics.
López-López O, Cerdán ME, González-Siso MI. López-López O, et al. Life (Basel). 2013 Apr 25;3(2):308-20. doi: 10.3390/life3020308. Life (Basel). 2013. PMID: 25369743 Free PMC article. Review. - Comparisons of clustered regularly interspaced short palindromic repeats and viromes in human saliva reveal bacterial adaptations to salivary viruses.
Pride DT, Salzman J, Relman DA. Pride DT, et al. Environ Microbiol. 2012 Sep;14(9):2564-76. doi: 10.1111/j.1462-2920.2012.02775.x. Epub 2012 May 15. Environ Microbiol. 2012. PMID: 22583485 Free PMC article. - CRISPR associated diversity within a population of Sulfolobus islandicus.
Held NL, Herrera A, Cadillo-Quiroz H, Whitaker RJ. Held NL, et al. PLoS One. 2010 Sep 28;5(9):e12988. doi: 10.1371/journal.pone.0012988. PLoS One. 2010. PMID: 20927396 Free PMC article.
References
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. - PubMed
- Mojica FJ, Diez-Villasenor C, Soria E, Juez G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol Microbiol. 2000;36:244–246. - PubMed
- Mojica FJ, Ferrer C, Juez G, Rodriguez-Valera F. Long stretches of short tandem repeats are present in the largest replicons of the Archaea Haloferax mediterranei and Haloferax volcanii and could be involved in replicon partitioning. Mol Microbiol. 1995;17:85–93. - PubMed
- Godde JS, Bickerton A. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J Mol Evol. 2006;62:718–729. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous