Functional reconstitution of ESCRT-III assembly and disassembly - PubMed (original) (raw)
Functional reconstitution of ESCRT-III assembly and disassembly
Suraj Saksena et al. Cell. 2009.
Abstract
Receptor downregulation in the MVB pathway is mediated by the ESCRT complexes. ESCRT-III is composed of four protein subunits that are monomeric in the cytosol and oligomerize into a protein lattice only upon membrane binding. Recent studies have shown that the ESCRT-III protein Snf7 can form a filament by undergoing homo-oligomerization. To examine the role of membrane binding and of interactions with other ESCRT components in initiating Snf7 oligomerization, we used fluorescence spectroscopy to directly detect and characterize the assembly of the Snf7 oligomer on liposomes using purified ESCRT components. The observed fluorescence changes reveal an obligatory sequence of membrane-protein and protein-protein interactions that generate the active conformation of Snf7. Also, we demonstrate that ESCRT-III assembly drives membrane deformation. Furthermore, using an in vitro disassembly assay, we directly demonstrate that Vps24 and Vps2 function as adaptors in the ATP-dependent membrane disassembly of the ESCRT-III complex by recruiting the AAA ATPase Vps4.
Figures
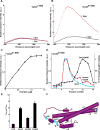
Fig. 1. Vps20 binding to membranes
Fluorescence emission spectra of 750 nM Vps207-NBD (A) and 1 μM Vps2061-NBD (B) before (black) and after (red) addition of excess (1.6 mM; see Fig. 1C) PC/PS/PI. (C) Titration of 1 μM Vps2061-NBD with liposomes reveals that fluorescence-detected (λem = 530 nm) binding is complete after addition of 1.2 mM PC/PS/PI. (D) Sepharose CL-2B chromatography of 800 nM Vps2061-NBD that had been preincubated with either 1.5 mM PC/PS/PI (red) or no liposomes (black). Protein was detected by NBD emission intensity and liposomes by light scattering (cyan). The elution profile of liposomes that were not exposed to protein was identical to the profile shown in this and later figures (data not shown). (E) The ratios of NBD emission intensities at 530 nm are shown for each derivative with (Fbound) and without (Ffree) membranes. (F) Cartoon representation of the water-soluble hVps24 monomer. Based on the sequence similarities of the ESCRT-III proteins, Vps20, Snf7 and Vps2 are believed to share structural features with the solved crystal structure of hVps24. Helices α1–5 and the N- and C-termini of the protein are labeled. Residues that were mutated to cysteine for labeling with NBD are depicted in cyan and labeled accordingly. Helix α6 as well as the α5–α6 loop have been manually sketched.
Fig. 2. Vps20 interactions with the Vps25 component of ESCRT-II
Fluorescence emission scans of 1 μM Vps20170-NBD D (A) and Vps2061-NBD (B) are shown in buffer (black) and after addition of 30 μM Vps25 (red). (C) The dependence of the emission intensities (F; λex = 468, λem = 530 nm) of 1 μM Vps20170-NBD (black) and Vps2061-NBD (red) on Vps25 addition are shown; F0 is the intensity in the absence of Vps25. (D) Emission scans of 1 μM Vps2061-NBD are shown in buffer (black), after addition of excess 1.5 mM liposomes (red), and after the addition of 30 μM Vps25 to Vps2061-NBD preincubated with 1.5 mM liposomes. (E) The dependence of the emission intensities (F; λex = 468, λem = 530 nm) of 1 μM Vps20170-NBD (black) and Vps2061-NBD (red) on Vps25 concentration is shown after the Vps20 derivatives were incubated with 1.5 mM liposomes for 30 min at 22 °C before Vps25 addition; F0 is the intensity in the absence of Vps25. (F) Summary of changes in the emission intensities of different NBD-labeled Vps20 mutants upon incubation with liposomes (red bar); Vps25 alone (dark blue bar); or liposomes and Vps25 (cyan). In each case, F0 represents emission intensity of the protein in buffer; and F represents emission intensity of the protein following incubation either with liposomes, Vps25 or liposomes and Vps25.
Fig. 3. Snf7 binding to membranes
Fluorescence emission spectra of 540 nM Snf781-NBD (A) and 300 nM Snf7200-NBD (B) before (black) and after (red) addition of an excess (1.5 mM; see Fig. 1C) of PC/PS/PI. (C) The ratios of NBD emission intensities at 530 nm are shown for each derivative with (Fbound) and without (Ffree) membranes. (D) Cartoon representation of the water-soluble hVps24 monomer as in Fig. 1F, but here showing the locations of the probes used in the Snf7 experiments (cyan). (E) Titration of 300 nM Snf7200-NBD with liposomes reveals that fluorescence-detected (λem = 530 nm) binding is complete after addition of 1.2 mM PC/PS/PI. (F) Sepharose CL-2B chromatography of 540 nM Snf781-NBD that had been preincubated with either 1.5 mM PC/PS/PI (red) or no liposomes (black). Protein was detected by NBD emission intensity, and liposomes by light scattering (cyan).
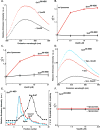
Fig. 4. Vps20 binding to membrane-bound Snf7
Fluorescence emission spectra of 300 nM Snf7200-NBD (A) and 2.2 μM Snf758-NBD (B) before (black) and after (red) addition of Vps20. Titration (λem = 530 nm) of 300 nM Snf7200-NBD (black) and 2.2 μM Snf758-NBD (red) with Vps20 either before (C) or after incubation with 1.5 mM PC/PS/PI (D). (E) Emission spectra of 2.2 μM Snf758-NBD bound to 1.5 mM PC/PS/PI before (red) and after (cyan) addition of 2.7 μM Vps20. For comparison, the spectrum of 2.2 μM Snf758-NBD in buffer is shown (black). (F) Sepharose CL-2B chromatography of samples containing 540 nM Snf781-NBD and 2.7 μM Vps20 that had been preincubated with either 1.5 mM PC/PS/PI (red) or no liposomes (black). Protein was detected by NBD emission intensity, and liposomes by light scattering (cyan).
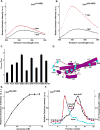
Fig. 5. Oligomerization of Snf7
(A) Emission spectrum (λex = 478 nm) of the DA sample (red) containing 320 nM Snf781-NBD, 4.2 μM of Snf781-Rh, 0.5 μM Vps20, and 1.5 mM PC/PS/PI. Unlabeled Snf781-C was substituted for one or both of the fluorescent derivatives to form the D (black), A (cyan), and B (magenta) samples. (B) The normalized net D (black) and net DA (red) emission spectra obtained from the samples in A. (C) Purified ESCRT-III proteins were mixed in different combinations and using different molar ratios (as indicated) in the presence or absence of PC/PS/PI. The liposomal fraction was isolated by centrifugation, detergent-solubilized, and analyzed by velocity sedimentation. (D) In wild-type (wt) cells, GFP-tagged carboxypeptidase-S (GFP-CPS) accumulates in the vacuolar lumen, while the FM4-64 dye stains the limiting membrane of the vacuole. In snf7Δ cells expressing the different mutant forms of Snf7 or in vps20Δ cells expressing the Vps20PW mutant, GFP-CPS co-localizes either with the limiting membrane of the vacuole or the class E compartment. (E) Endosome (P13) fractions prepared from vps4Δsnf7Δ cells expressing either wt Snf7 (top panel) or the different Snf7 mutants were detergent solubilized, and analyzed using glycerol velocity gradients.
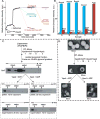
Fig. 6. ESCRT-III assembly, disassembly and liposome deformation
(A) Time- and component-dependent emission intensity profiles. Each reaction sample (A–F) contained 540 nM Snf781-NBD mixed with Vps20 at time 0. At 600 sec, 1.5mM PC/PS/PI was added to each sample. At 1200 sec, additions were made as follows: sample A (orange trace) to 180 nM Vps2, 1 μM Vps4, 1mM ATP; B (magenta) to 270 nM Vps24, 1 μM Vps4, 1mM ATP; C (cyan) to 270 nM Vps24, 180 nM Vps2, 1 μM Vps4 (E233Q), 1mM ATP; D (green) to 270 nM Vps24, 180 nM Vps2, 1 μM Vps4, 1mM ADP; E (red) to 270 nM Vps24, 180 nM Vps2, 1 μM Vps4, 1mM ATP; and F (purple) received only buffer. After incubation (37°C, 45 min), emission was re-measured from 3900–4500 sec at 22°C. Samples A, B, and F then received 270 nM Vps24, 180 nM Vps2, or buffer, respectively, were incubated (37°C, 45 min), and emission was re-measured from 7200–7800 sec. (B) In some experiments, samples were analyzed by Sepharose CL-2B gel filtration chromatography after completing the first 45 min incubation. Protein was detected by NBD emission intensity, and liposomes by light scattering (cyan). The red and blue bars indicate the relative amounts of free Snf781-NBD and liposome-bound Snf781-NBD respectively. (C) Outline of the in vitro ESCRT-III disassembly assay. ESCRT-III complex was assembled on liposomes using purified ESCRT-III proteins and centrifuged to separate the liposome-bound complex The liposome-bound ESCRT-III complex was treated with either (i) Vps4 and ATP or (ii) Vps4 and ADP for 45 min at 37°C. Following incubation, the samples were centrifuged to separate the liposome-bound proteins (“pellet”) from free ESCRT-III proteins (“supernatant”), and then further analyzed by velocity sedimentation. (D) Negative stain EM analyses of PC/PS/PI liposomes, ESCRT-III bound liposomes, ESCRT-III-bound liposomes treated with 1μM Vps4 + 1mM ATP, and ESCRT-III-bound liposomes treated with 1 μM Vps4 + 1mM ADP.
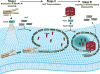
Fig. 7. Speculative model for the ESCRT-III reaction cycle
ESCRT-III assembly is initiated when Vps20 binds to Vps25 on the membrane surface. In the second stage, Snf7 binds to the membrane-bound Vps25-Vps20 complex, and this triggers the formation of a membrane-bound Snf7 oligomer. The Snf7 oligomer traps cargo into a localized sorting domain and induces membrane deformation. Vps24 caps the Snf7 oligomer, and interactions between Vps2 and Vps4 initiate membrane disassembly of the ESCRT-III complex (see Discussion).
Comment in
- ESCRTing membrane deformation.
Malerød L, Stenmark H. Malerød L, et al. Cell. 2009 Jan 9;136(1):15-7. doi: 10.1016/j.cell.2008.12.029. Cell. 2009. PMID: 19135881
Similar articles
- Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation.
Teis D, Saksena S, Emr SD. Teis D, et al. Dev Cell. 2008 Oct;15(4):578-89. doi: 10.1016/j.devcel.2008.08.013. Dev Cell. 2008. PMID: 18854142 - Membrane scission by the ESCRT-III complex.
Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Wollert T, et al. Nature. 2009 Mar 12;458(7235):172-7. doi: 10.1038/nature07836. Epub 2009 Feb 22. Nature. 2009. PMID: 19234443 Free PMC article. - Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24.
Ghazi-Tabatabai S, Saksena S, Short JM, Pobbati AV, Veprintsev DB, Crowther RA, Emr SD, Egelman EH, Williams RL. Ghazi-Tabatabai S, et al. Structure. 2008 Sep 10;16(9):1345-56. doi: 10.1016/j.str.2008.06.010. Structure. 2008. PMID: 18786397 - Structures, Functions, and Dynamics of ESCRT-III/Vps4 Membrane Remodeling and Fission Complexes.
McCullough J, Frost A, Sundquist WI. McCullough J, et al. Annu Rev Cell Dev Biol. 2018 Oct 6;34:85-109. doi: 10.1146/annurev-cellbio-100616-060600. Epub 2018 Aug 10. Annu Rev Cell Dev Biol. 2018. PMID: 30095293 Free PMC article. Review. - ESCRT-III and Vps4: a dynamic multipurpose tool for membrane budding and scission.
Alonso Y Adell M, Migliano SM, Teis D. Alonso Y Adell M, et al. FEBS J. 2016 Sep;283(18):3288-302. doi: 10.1111/febs.13688. Epub 2016 Mar 8. FEBS J. 2016. PMID: 26910595 Review.
Cited by
- An expanded view of the eukaryotic cytoskeleton.
Moseley JB. Moseley JB. Mol Biol Cell. 2013 Jun;24(11):1615-8. doi: 10.1091/mbc.E12-10-0732. Mol Biol Cell. 2013. PMID: 23722945 Free PMC article. - ESCRT-III CHMP2A and CHMP3 form variable helical polymers in vitro and act synergistically during HIV-1 budding.
Effantin G, Dordor A, Sandrin V, Martinelli N, Sundquist WI, Schoehn G, Weissenhorn W. Effantin G, et al. Cell Microbiol. 2013 Feb;15(2):213-26. doi: 10.1111/cmi.12041. Epub 2012 Nov 6. Cell Microbiol. 2013. PMID: 23051622 Free PMC article. - A unique role for the host ESCRT proteins in replication of Tomato bushy stunt virus.
Barajas D, Jiang Y, Nagy PD. Barajas D, et al. PLoS Pathog. 2009 Dec;5(12):e1000705. doi: 10.1371/journal.ppat.1000705. Epub 2009 Dec 24. PLoS Pathog. 2009. PMID: 20041173 Free PMC article. - Vps60 initiates alternative ESCRT-III filaments.
Pfitzner AK, Zivkovic H, Bernat-Silvestre C, West M, Peltier T, Humbert F, Odorizzi G, Roux A. Pfitzner AK, et al. J Cell Biol. 2023 Nov 6;222(11):e202206028. doi: 10.1083/jcb.202206028. Epub 2023 Sep 28. J Cell Biol. 2023. PMID: 37768378 Free PMC article. - SAXS ensemble refinement of ESCRT-III CHMP3 conformational transitions.
Różycki B, Kim YC, Hummer G. Różycki B, et al. Structure. 2011 Jan 12;19(1):109-16. doi: 10.1016/j.str.2010.10.006. Structure. 2011. PMID: 21220121 Free PMC article.
References
- Ghazi-Tabatabai S, Saksena S, Short JM, Pobbati AV, Veprintsev DB, Crowther RA, Emr SD, Egelman EH, Williams RL. Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure. 2008;16:1345–1356. - PubMed
- Johnson AE. Fluorescence approaches for determining protein conformations, interactions and mechanisms at membranes. Traffic. 2005;6:1078–1092. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases