Cooperative activation of the ATR checkpoint kinase by TopBP1 and damaged DNA - PubMed (original) (raw)
Cooperative activation of the ATR checkpoint kinase by TopBP1 and damaged DNA
Jun-Hyuk Choi et al. Nucleic Acids Res. 2009 Apr.
Abstract
TopBP1, acting in concert with DNA containing bulky base lesions, stimulates ATR kinase activity under physiologically relevant reaction conditions. Here, we analyze the roles of the three components in ATR activation: DNA, base damage and TopBP1. We show that base adducts caused by a potent carcinogen, benzo[a]pyrene diol epoxide (BPDE), constitute a strong signal for TopBP1-dependent ATR kinase activity on Chk1 and p53. We find that the C-terminus of TopBP1 binds preferentially to damaged DNA and is sufficient to mediate damaged DNA-dependent ATR activation in a manner similar to full-length TopBP1. Significantly, we find that stimulation of ATR by BPDE-damaged DNA exhibits strong dependence on the length of DNA, with essentially no stimulation with fragments of 0.2 kb and reaching maximum stimulation with 2 kb fragments. Moreover, TopBP1 shows preferential binding to longer DNA fragments and, in contrast to previous biochemical studies, TopBP1 binding is completely independent of DNA ends. We find that TopBP1 binds to circular and linear DNAs with comparable affinities and that these DNA forms elicit the same level of TopBP1-dependent ATR activation. Taken together, these findings suggest a cooperative activation mechanism for the ATR checkpoint kinase by TopBP1 and damaged DNA.
Figures
Figure 1.
TopBP1 fragments used in this study and their DNA binding properties. (A) Schematic of human TopBP1 and its fragments that were purified for structure–function experiments. The amino acid positions are indicated, and the eight boxes indicate the BRCT motifs. The ATR activating domain between BRCT domains 6 and 7 is indicated. (B) The GST-fusion proteins visualized by SDS–PAGE followed by Coomassie blue staining. (C) Preferential binding of TopBP1 and TopBP1 fragments to BPDE-damaged DNA. (Top panel) Unmodified (UM) or BPDE treated pUC19 plasmid DNAs (1 ng) which had been labeled with [γ-32P]ATP were incubated with 3 pmol of full-length TopBP1 (FL) bound beads or beads carrying 3 pmol of the TopBP1-A, -B, -C or -D fragments in buffer containing 200 mM NaCl. The bound DNA was eluted by proteinase K, analyzed by agarose gel electrophoresis, and visualized by autoradiography. The input lanes contain fifty percent of DNA added to the reaction. The results from two experiments were quantified and plotted.
Figure 2.
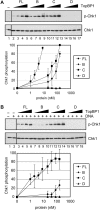
Stimulation of the ATR kinase by TopBP1 and its fragments. (A) TopBP1 fragments containing the ATR-activating domain stimulate ATR in the absence of DNA. ATR (0.4 nM) was incubated with Chk1 (10 nM) in the presence of full-length TopBP1 (1–8 nM) or the indicated TopBP1 fragments (20–160nM) under low ionic strength conditions (45 mM total salt concentration). ATR kinase activity was determined by immunoblotting for phospho-Chk1 (S345) and Chk1 as indicated. The graph shows quantitative analysis of the data from three independent experiments conducted under identical conditions. (B) DNA stimulates the kinase activity of ATR in the presence of full-length TopBP1 or the C-terminal fragment. Kinase assays were performed as described in Figure 2A, except with 5 ng of BPDE-damaged DNA under high ionic strength conditions (85 mM total salt concentration). The average levels of Chk1 phosphorylation from three independent experiments were quantified and graphed.
Figure 3.
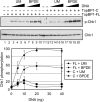
TopBP1 C-terminal fragment stimulates the ATR kinase in a manner dependent on the presence of damaged DNA. Kinase assays were carried out with ATR (0.4 nM), Chk1 (10 nM) and unmodified (UM) or BPDE-damaged DNA (5–40 ng) in the presence of full-length TopBP1 (0.5 nM) or TopBP1 C-fragment (10 nM) under high ionic strength conditions. The average levels of Chk1 phosphorylation from three independent experiments were quantified and graphed.
Figure 4.
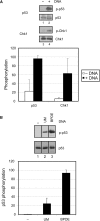
Damaged DNA-dependent stimulation of ATR is independent of the substrate. (A) Addition of DNA stimulates the ATR kinase activity toward another key downstream target, p53 in the presence of TopBP1. Kinase assays were carried out with ATR and TopBP1-C fragment in the absence or presence of 5 ng of unmodified circular DNA in reactions containing 5 nM Chk1 (lane 1 and 2) or 5 nM p53 (lane 3 and 4) as described in Figure 3. The average levels of Chk1 and p53 phosphorylation from three independent experiments were quantified and graphed. (B) Phosphorylation of p53 is strongly stimulated by BPDE-damaged DNA. Kinase assays were performed with ATR, p53 and 5ng of unmodified (UM) (lane 2) or BPDE-damaged DNA (BPDE) (lane 3) in the presence of full-length TopBP1 as described in Figure 3. The average levels of p53 phosphorylation from two independent experiments were quantified and graphed.
Figure 5.
Efficient ATR activation and DNA binding by TopBP1 depend on the length of DNA. (A) TopBP1-dependent ATR stimulation increases with DNA length. Kinase assays were performed as described in Figure 3, except with 1.25, 2.5, and 5 ng of DNAs of different lengths ranging from 0.2 to 2.6 kb. The average levels of Chk1 phosphorylation from three independent experiments were quantified and graphed. (B) TopBP1 preferentially binds to longer DNA. DNA-binding assays were carried out as described in Figure 1C, except with equal molar or mass amounts of 0.2 or 2.6 kb DNA fragments in buffer containing 300 mM NaCl. The results from two experiments were quantified and plotted. (C) TopBP1 preferentially binds longer DNAs under kinase reaction conditions. DNA-binding assays were performed with 0.15 pmol of TopBP1 and 3.5 fmol of 0.2 or 2.6 kb DNA fragments under conditions used for kinase reactions. The results from two experiments were quantified and graphed.
Figure 6.
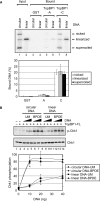
DNA binding and ATR stimulatory activities of TopBP1 are independent of DNA ends. (A) TopBP1 has no preferential binding to DNA ends. Covalently closed circular and linear DNA that had been labeled with [γ-32P]ATP were incubated with GST alone, the TopBP1-A or TopBP1-C fragment immobilized on glutathione beads. After extensive washing, the bound DNA was eluted and analyzed on an agarose gel containing ethidium bromide. The gel was dried and visualized by autoradiography. Note that the ‘covalently closed DNA’ sample contains covalently closed, nicked and linear DNAs and hence gives a fair representation of the relative affinities of these three forms to the TopBP1 fragments. The input lanes contain fifty percent of DNA added to the reaction. The results from five experiments were quantified and plotted. (B) DNA double-strand breaks has no effect on TopBP1-dependent ATR activation. Kinase assays were carried out with ATR, Chk1 and TopBP1 in the presence of unmodified (UM) or BPDE-damaged circular or linear DNA as described in Figure 3. The average levels of Chk1 phosphorylation from three independent experiments were quantified and graphed.
Similar articles
- Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation.
Liu S, Bekker-Jensen S, Mailand N, Lukas C, Bartek J, Lukas J. Liu S, et al. Mol Cell Biol. 2006 Aug;26(16):6056-64. doi: 10.1128/MCB.00492-06. Mol Cell Biol. 2006. PMID: 16880517 Free PMC article. - Reconstitution of a human ATR-mediated checkpoint response to damaged DNA.
Choi JH, Lindsey-Boltz LA, Sancar A. Choi JH, et al. Proc Natl Acad Sci U S A. 2007 Aug 14;104(33):13301-6. doi: 10.1073/pnas.0706013104. Epub 2007 Aug 8. Proc Natl Acad Sci U S A. 2007. PMID: 17686975 Free PMC article. - Activation of ATR-related protein kinase upon DNA damage recognition.
Ma M, Rodriguez A, Sugimoto K. Ma M, et al. Curr Genet. 2020 Apr;66(2):327-333. doi: 10.1007/s00294-019-01039-w. Epub 2019 Oct 17. Curr Genet. 2020. PMID: 31624858 Free PMC article. Review.
Cited by
- The effect of replication protein A inhibition and post-translational modification on ATR kinase signaling.
Jordan MR, Oakley GG, Mayo LD, Balakrishnan L, Turchi JJ. Jordan MR, et al. Sci Rep. 2024 Aug 26;14(1):19791. doi: 10.1038/s41598-024-70589-y. Sci Rep. 2024. PMID: 39187637 Free PMC article. - Multi-BRCT scaffolds use distinct strategies to support genome maintenance.
Wan B, Hang LE, Zhao X. Wan B, et al. Cell Cycle. 2016 Oct;15(19):2561-2570. doi: 10.1080/15384101.2016.1218102. Epub 2016 Aug 11. Cell Cycle. 2016. PMID: 27580271 Free PMC article. Review. - Interactions of human mismatch repair proteins MutSalpha and MutLalpha with proteins of the ATR-Chk1 pathway.
Liu Y, Fang Y, Shao H, Lindsey-Boltz L, Sancar A, Modrich P. Liu Y, et al. J Biol Chem. 2010 Feb 19;285(8):5974-82. doi: 10.1074/jbc.M109.076109. Epub 2009 Dec 22. J Biol Chem. 2010. PMID: 20029092 Free PMC article. - ATR Kinase Inhibition Protects Non-cycling Cells from the Lethal Effects of DNA Damage and Transcription Stress.
Kemp MG, Sancar A. Kemp MG, et al. J Biol Chem. 2016 Apr 22;291(17):9330-42. doi: 10.1074/jbc.M116.719740. Epub 2016 Mar 3. J Biol Chem. 2016. PMID: 26940878 Free PMC article.
References
- Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. - PubMed
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. - PubMed
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 2006;8:37–45. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous