Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs - PubMed (original) (raw)
Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs
Fernando Cruz-Guilloty et al. J Exp Med. 2009.
Abstract
Activation of naive CD8(+) T cells with antigen induces their differentiation into effector cytolytic T lymphocytes (CTLs). CTLs lyse infected or aberrant target cells by exocytosis of lytic granules containing the pore-forming protein perforin and a family of proteases termed granzymes. We show that effector CTL differentiation occurs in two sequential phases in vitro, characterized by early induction of T-bet and late induction of Eomesodermin (Eomes), T-box transcription factors that regulate the early and late phases of interferon (IFN) gamma expression, respectively. In addition, we demonstrate a critical role for the transcription factor Runx3 in CTL differentiation. Runx3 regulates Eomes expression as well as expression of three cardinal markers of the effector CTL program: IFN-gamma, perforin, and granzyme B. Our data point to the existence of an elaborate transcriptional network in which Runx3 initially induces and then cooperates with T-box transcription factors to regulate gene transcription in differentiating CTLs.
Figures
Figure 1.
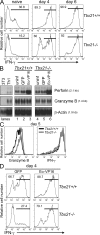
Kinetics of gene expression during CD8+ T cell differentiation. (A) Kinetics of Prf1, Gzmb, Tbx21 (T-bet), and Eomes mRNA expression in differentiating P14 CD8+ T cells analyzed by Northern blotting. RNA from day 7 Th1 cells was used as a control. Sizes of mRNA transcripts are indicated. (B) Quantification of relative mRNA amounts by phosphorimager analysis. (C) Kinetics of protein expression in differentiating P14 CD8+ T cells analyzed by immunoblotting. Sizes of protein bands are indicated. (D) Relative protein amounts quantified from the Western blots. (E) Intracellular staining for granzyme B, IFN-γ, and TNF. Granzyme B staining was specific relative to an isotype control (not depicted). Cells were restimulated with PMA and ionomycin for 4 h. (F) FACS-based assay to measure cytolytic activity of P14 CD8+ T cells against EL4 targets loaded with 0 (−) or 1 (+) μM Gp33 peptide (effector-to-target ratio = 5:1). Percentage of Annexin V+ (apoptotic) target cells in the CD8-negative EL4 target population (dot plots) was determined (histograms). Cytolytic activity was blocked by incubation with 2 mM EGTA (not depicted), confirming involvement of the granule exocytosis (perforin–granzyme B) pathway. Data are representative of at least five (A–E) or three (F) independent experiments.
Figure 2.
Regulation of perforin, granzyme B, and IFN-γ expression by T-bet and Eomes in differentiating CTLs. (A) IFN-γ expression by WT (Tbx21+/+) and T-bet–deficient (Tbx21−/−) T cells. Naive CD8+ T cells, or cells activated and cultured for 4 or 6 d, were restimulated with PMA and ionomycin for 6 h, and IFN-γ expression was assessed by intracellular staining. Numbers show the percentage of IFN-γ+ cells. (B) Northern blot analysis of Prf1 and GzmB mRNA expression in WT or T-bet–deficient CD8+ T cells activated and either left uninfected (uninf) or transduced with retroviruses expressing Eomes-VP16 (Eo-VP16) or an empty IRES-GFP cassette (GFP). Total cellular RNA was analyzed on day 6 of culture. The frequency of transduced cells in the cultures was equivalent for both constructs (∼65–70% GFP+ cells; not depicted). (C) Granzyme B and IFN-γ expression by Tbx21+/+ and Tbx21−/− T cells analyzed in restimulated cells that had been cultured for 5 d. (D) IFN-γ production by cells transduced with Eo-VP16 or control (GFP) retroviruses (RV) measured on day 4 after 6 h of restimulation with PMA and ionomycin. Numbers show the percentage of GFP+ IFN-γ+ cells. Results are representative of three (A and C) or two (B and D) independent experiments.
Figure 3.
Key role for Runx3 in effector CTL differentiation. (A) Western analysis of Runx3, Eomes, T-bet, and perforin expression in Runx3+/+ versus Runx3−/− CD8+ SP T cells differentiated for 6 d. β-Actin was used as a loading control. (B) Northern blot analysis of Prf1 mRNA expression in Runx3+/+ versus Runx3−/− CD8+ T cells differentiated for 6 d. β–Actin was used as a loading control. (C) Expression of granzyme B, IFN-γ, TNF, and IL-2 by resting or restimulated (6 h) Runx3+/+ versus Runx3−/− CD8+ SP T cells differentiated for 6 d. The vertical gray line indicates the granzyme B MFI for WT GFP+ cells. Results in A–C are representative of two independent experiments. (D) ChIP analysis of binding of endogenous Runx3 and Eomes to the Prf1 locus. Enrichment of the indicated genomic regions was evaluated by real-time PCR of DNA from immunoprecipitated and input chromatin. The data are the means of duplicate measurements from two chromatin preparations from two independent CD8+ T cell differentiations. The efficiency of recovery of input for the −1-kb region of Prf1 was 0.97% for the Runx3 ChIP and 0.5% for the Eomes ChIP.
Figure 4.
Runx3 controls Eomes, perforin, granzyme B, and IFN-γ expression in effector CTLs. Runx3+/+ or Runx3−/− CD8+ T cells were activated and transduced with retroviruses bearing an empty IRES-GFP cassette (GFP) or also encoding Eomes-VP16 (Eo-VP16) or Myc-Runx3 (Runx3). The frequency of transduced cells in the cultures was equivalent for all constructs (∼75–90% GFP+ cells; not depicted). (A) Protein expression in whole-cell extracts (day 6) was analyzed by immunoblotting. Overexpression of Eomes-VP16 cannot be detected with the Eomes antibody, as the C-terminal epitope is within the region that has been replaced with the VP16 transactivation domain. (B) Expression of granzyme B and IFN-γ after culture for 6 d and restimulation for 4 h with PMA and ionomycin was determined by intracellular staining. The percentage of positively stained cells is shown above the gate; the mean fluorescence intensity (MFI) of granzyme B staining for the total population is shown below the gate. The vertical gray lines indicate the MFI for WT GFP+ cells. Results are representative of at least two independent experiments. (C) Schematic diagram of the transcriptional network involving Runx3 and T-box factors. T-bet is induced by TCR signals and is essential for early IFN-γ expression. Runx3 is present in naive CD8+ T cells and represses Runx1 and induces Eomes, perforin, granzyme B, and IFN-γ expression. Eomes may participate in sustaining late IFN-γ expression, whereas Runx3 and Eomes (but not T-bet) may cooperate to activate perforin expression. The dotted line indicates the partial effect of T-bet deficiency on Gzmb mRNA but not granzyme B protein expression.
Similar articles
- Notch regulates cytolytic effector function in CD8+ T cells.
Cho OH, Shin HM, Miele L, Golde TE, Fauq A, Minter LM, Osborne BA. Cho OH, et al. J Immunol. 2009 Mar 15;182(6):3380-9. doi: 10.4049/jimmunol.0802598. J Immunol. 2009. PMID: 19265115 Free PMC article. - Natural Killer Cell Group 7 Sequence in Cytotoxic Cells Optimizes Exocytosis of Lytic Granules Essential for the Perforin-Dependent, but Not Fas Ligand-Dependent, Cytolytic Pathway.
Morikawa Y, Murakami M, Kondo H, Nemoto N, Iwabuchi K, Eshima K. Morikawa Y, et al. Immunohorizons. 2021 Apr 28;5(4):234-245. doi: 10.4049/immunohorizons.2100029. Immunohorizons. 2021. PMID: 33911019 - Ectopic expression of a T-box transcription factor, eomesodermin, renders CD4(+) Th cells cytotoxic by activating both perforin- and FasL-pathways.
Eshima K, Chiba S, Suzuki H, Kokubo K, Kobayashi H, Iizuka M, Iwabuchi K, Shinohara N. Eshima K, et al. Immunol Lett. 2012 May 30;144(1-2):7-15. doi: 10.1016/j.imlet.2012.02.013. Epub 2012 Mar 10. Immunol Lett. 2012. PMID: 22425747 - The transcriptional control of the perforin locus.
Pipkin ME, Rao A, Lichtenheld MG. Pipkin ME, et al. Immunol Rev. 2010 May;235(1):55-72. doi: 10.1111/j.0105-2896.2010.00905.x. Immunol Rev. 2010. PMID: 20536555 Free PMC article. Review. - Recent developments in the transcriptional regulation of cytolytic effector cells.
Glimcher LH, Townsend MJ, Sullivan BM, Lord GM. Glimcher LH, et al. Nat Rev Immunol. 2004 Nov;4(11):900-11. doi: 10.1038/nri1490. Nat Rev Immunol. 2004. PMID: 15516969 Review.
Cited by
- The Contradictory Role of Interleukin-33 in Immune Cells and Tumor Immunity.
Zhang X, Chen W, Zeng P, Xu J, Diao H. Zhang X, et al. Cancer Manag Res. 2020 Aug 21;12:7527-7537. doi: 10.2147/CMAR.S262745. eCollection 2020. Cancer Manag Res. 2020. PMID: 32904627 Free PMC article. Review. - Role of nuclear localization in the regulation and function of T-bet and Eomes in exhausted CD8 T cells.
McLane LM, Ngiow SF, Chen Z, Attanasio J, Manne S, Ruthel G, Wu JE, Staupe RP, Xu W, Amaravadi RK, Xu X, Karakousis GC, Mitchell TC, Schuchter LM, Huang AC, Freedman BD, Betts MR, Wherry EJ. McLane LM, et al. Cell Rep. 2021 May 11;35(6):109120. doi: 10.1016/j.celrep.2021.109120. Cell Rep. 2021. PMID: 33979613 Free PMC article. - CD8(+) T-cell effector function and transcriptional regulation during HIV pathogenesis.
Demers KR, Reuter MA, Betts MR. Demers KR, et al. Immunol Rev. 2013 Jul;254(1):190-206. doi: 10.1111/imr.12069. Immunol Rev. 2013. PMID: 23772621 Free PMC article. Review. - Regulation of CD4 T Cell Responses by the Transcription Factor Eomesodermin.
Dhume K, Kaye B, McKinstry KK. Dhume K, et al. Biomolecules. 2022 Oct 24;12(11):1549. doi: 10.3390/biom12111549. Biomolecules. 2022. PMID: 36358898 Free PMC article. Review. - Biology and clinical relevance of follicular cytotoxic T cells.
Lv Y, Ricard L, Gaugler B, Huang H, Ye Y. Lv Y, et al. Front Immunol. 2022 Dec 14;13:1036616. doi: 10.3389/fimmu.2022.1036616. eCollection 2022. Front Immunol. 2022. PMID: 36591286 Free PMC article. Review.
References
- Harty, J.T., A.R. Tvinnereim, and D.W. White. 2000. CD8+ T cell effector mechanisms in resistance to infection. Annu. Rev. Immunol. 18:275–308. - PubMed
- Williams, M.A., and M.J. Bevan. 2007. Effector and memory CTL differentiation. Annu. Rev. Immunol. 25:171–192. - PubMed
- Badovinac, V.P., and J.T. Harty. 2006. Programming, demarcating, and manipulating CD8+ T-cell memory. Immunol. Rev. 211:67–80. - PubMed
- Ansel, K.M., I. Djuretic, B. Tanasa, and A. Rao. 2006. Regulation of Th2 differentiation and Il4 locus accessibility. Annu. Rev. Immunol. 24:607–656. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 CA126247/CA/NCI NIH HHS/United States
- R01 AI044432/AI/NIAID NIH HHS/United States
- P30 AI073961/AI/NIAID NIH HHS/United States
- R37 CA042471/CA/NCI NIH HHS/United States
- F32 CA126247-01/CA/NCI NIH HHS/United States
- CA42471/CA/NCI NIH HHS/United States
- R01 CA042471/CA/NCI NIH HHS/United States
- AI44432/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials