Optimization of gene expression by natural selection - PubMed (original) (raw)
Optimization of gene expression by natural selection
Trevor Bedford et al. Proc Natl Acad Sci U S A. 2009.
Abstract
It is generally assumed that stabilizing selection promoting a phenotypic optimum acts to shape variation in quantitative traits across individuals and species. Although gene expression represents an intensively studied molecular phenotype, the extent to which stabilizing selection limits divergence in gene expression remains contentious. In this study, we present a theoretical framework for the study of stabilizing and directional selection using data from between-species divergence of continuous traits. This framework, based upon Brownian motion, is analytically tractable and can be used in maximum-likelihood or Bayesian parameter estimation. We apply this model to gene-expression levels in 7 species of Drosophila, and find that gene-expression divergence is substantially curtailed by stabilizing selection. However, we estimate the selective effect, s, of gene-expression change to be very small, approximately equal to Ns for a change of one standard deviation, where N is the effective population size. These findings highlight the power of natural selection to shape phenotype, even when the fitness effects of mutations are in the nearly neutral range.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
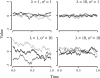
Fig. 1.
Realizations of the OU process. Three individual realizations are shown for each of four different parameter values. The drift parameter σ determines the degree of mutational pressure randomly impacting the trait value, while λ determines the pull of selection toward some optimal trait value (in this case 0). In each realization, the starting value was sampled from the equilibrium distribution.
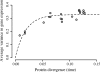
Fig. 2.
Average pairwise variance in expression level for Drosophila species. Each point represents the average variance between a species pair. This variance initially increases with time, but eventually saturates. In the absence of stabilizing selection, pairwise variance is expected to saturate at 1. Nonlinear regression fit of pairwise variance vs. time for the OU model is represented as a dashed line (λ = 26.14; σ = 4.14).
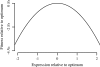
Fig. 3.
Fitness landscape of gene-expression level estimated from OU parameters. Expression level is measured in terms of standard deviations relative to other genes in the genome. Fitness is equal to −(λ/2σ2)(μ−z)2, where z represents the current trait value. The quadratic shape of the fitness landscape is assumed by the OU model; the data provides the magnitude of curvature.
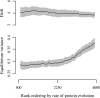
Fig. 4.
Effect of protein-sequence evolution on patterns of gene-expression divergence. Nonlinear regression was used to estimate the drift parameter σ and equilibrium variance σ2/2λ in sliding windows across gene rank ordered according to their rate of protein-sequence evolution. Each window consists of 1,125 genes, or 25% of the total set of genes in which reliable alignments could be made. Mean estimates are shown as solid lines and 95% CIs shown as gray boundaries. Fast-evolving genes show similar rates of drift, but significantly greater levels of equilibrium variance, compared to slow-evolving genes.
Similar articles
- Common pattern of evolution of gene expression level and protein sequence in Drosophila.
Nuzhdin SV, Wayne ML, Harmon KL, McIntyre LM. Nuzhdin SV, et al. Mol Biol Evol. 2004 Jul;21(7):1308-17. doi: 10.1093/molbev/msh128. Epub 2004 Mar 19. Mol Biol Evol. 2004. PMID: 15034135 - Detecting selection with a genetic cross.
Fraser HB. Fraser HB. Proc Natl Acad Sci U S A. 2020 Sep 8;117(36):22323-22330. doi: 10.1073/pnas.2014277117. Epub 2020 Aug 26. Proc Natl Acad Sci U S A. 2020. PMID: 32848059 Free PMC article. - Causes of natural variation in fitness: evidence from studies of Drosophila populations.
Charlesworth B. Charlesworth B. Proc Natl Acad Sci U S A. 2015 Feb 10;112(6):1662-9. doi: 10.1073/pnas.1423275112. Epub 2015 Jan 8. Proc Natl Acad Sci U S A. 2015. PMID: 25572964 Free PMC article. Review. - Experimental evolution with Drosophila.
Burke MK, Rose MR. Burke MK, et al. Am J Physiol Regul Integr Comp Physiol. 2009 Jun;296(6):R1847-54. doi: 10.1152/ajpregu.90551.2008. Epub 2009 Apr 1. Am J Physiol Regul Integr Comp Physiol. 2009. PMID: 19339679 Review.
Cited by
- Multi-omic analysis of signalling factors in inflammatory comorbidities.
Xiao H, Bartoszek K, Lio' P. Xiao H, et al. BMC Bioinformatics. 2018 Nov 30;19(Suppl 15):439. doi: 10.1186/s12859-018-2413-x. BMC Bioinformatics. 2018. PMID: 30497370 Free PMC article. - Inferring evolutionary histories of pathway regulation from transcriptional profiling data.
Schraiber JG, Mostovoy Y, Hsu TY, Brem RB. Schraiber JG, et al. PLoS Comput Biol. 2013;9(10):e1003255. doi: 10.1371/journal.pcbi.1003255. Epub 2013 Oct 10. PLoS Comput Biol. 2013. PMID: 24130471 Free PMC article. - Expression of defense genes in Drosophila evolves under a different selective regime from expression of other genes.
Wayne ML, Pienaar J, Telonis-Scott M, Sylvestre LS, Nuzhdin SV, McIntyre LM. Wayne ML, et al. Evolution. 2011 Apr;65(4):1068-78. doi: 10.1111/j.1558-5646.2010.01197.x. Epub 2010 Dec 22. Evolution. 2011. PMID: 21108635 Free PMC article. - Regulatory Divergence as a Mechanism for X-Autosome Incompatibilities in Caenorhabditis Nematodes.
Viswanath A, Cutter AD. Viswanath A, et al. Genome Biol Evol. 2023 Apr 6;15(4):evad055. doi: 10.1093/gbe/evad055. Genome Biol Evol. 2023. PMID: 37014784 Free PMC article. - A selection index for gene expression evolution and its application to the divergence between humans and chimpanzees.
Warnefors M, Eyre-Walker A. Warnefors M, et al. PLoS One. 2012;7(4):e34935. doi: 10.1371/journal.pone.0034935. Epub 2012 Apr 18. PLoS One. 2012. PMID: 22529958 Free PMC article.
References
- Smith NG, Eyre-Walker A. Adaptive protein evolution in Drosophila. Nature. 2002;415:1022–1024. - PubMed
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. - PubMed
- Carroll SB, Grenier JK, Weatherbee SD. From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design. New York: Blackwell; 2001.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous