Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells - PubMed (original) (raw)
. 2009 Jan 16;30(1):155-67.
doi: 10.1016/j.immuni.2008.12.009.
Lai Wei, Jinfang Zhu, Chongzhi Zang, Jane Hu-Li, Zhengju Yao, Kairong Cui, Yuka Kanno, Tae-Young Roh, Wendy T Watford, Dustin E Schones, Weiqun Peng, Hong-Wei Sun, William E Paul, John J O'Shea, Keji Zhao
Affiliations
- PMID: 19144320
- PMCID: PMC2722509
- DOI: 10.1016/j.immuni.2008.12.009
Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells
Gang Wei et al. Immunity. 2009.
Abstract
Multipotential naive CD4(+) T cells differentiate into distinct lineages including T helper 1 (Th1), Th2, Th17, and inducible T regulatory (iTreg) cells. The remarkable diversity of CD4(+) T cells begs the question whether the observed changes reflect terminal differentiation with heritable epigenetic modifications or plasticity in T cell responses. We generated genome-wide histone H3 lysine 4 (H3K4) and lysine 27 (H3K27) trimethylation maps in naive, Th1, Th2, Th17, iTreg, and natural Treg (nTreg) cells. We found that although modifications of signature-cytokine genes (Ifng, Il4, and Il17) partially conform to the expectation of lineage commitment, genes encoding transcription factors like Tbx21 exhibit a broad spectrum of epigenetic states, consistent with our demonstration of T-bet and interferon-gamma induction in nTreg cells. Our data suggest an epigenetic mechanism underlying the specificity and plasticity of effector and regulatory T cells and also provide a framework for understanding complexity of CD4(+) T helper cell differentiation.
Figures
Figure 1. Overview of the experimental design
Splenic and lymph node CD4+ T cells from C57BL/6J mice were sorted for CD4+CD62L+CD44-CD25- naïve population and CD4+CD25hi natural T regulatory cells, or cultured under Th1, Th2, Th17, and iTreg cell conditions. The genome-wide distribution of H3K4me3 and H3K27me3 was determined using ChIP-seq. Gene expression profiling of the six T cell subsets was performed using DNA microarrays.
Figure 2. Distribution of H3K4me3 and H3K27me3 islands
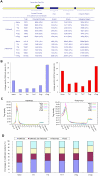
(A) The mouse genome was divided into four kinds of regions: proximal promoter (1 kb upstream and downstream of transcription start site), exon, intron, and intergenic regions. The number of islands for each sample was listed as the total identified islands (Total), followed by the islands among genomic regions with the percentage listed in the parenthesis. (B) Lineage-specific islands were identified as described in Supplemental Information. The percentage of lineage-specific H3K4me3 (left) and H3K27m3 (right) islands is expressed as a proportion of all islands identified in each lineage. (C) For each gene, uniquely mapped tags were summed in 1 kb windows for the regions from 5 kb upstream of the transcription start site (txStart) to the txStart and from the transcription end site (txEnd) to 5 kb downstream. Within the gene bodies, tags were summed in windows equal to 5% of the gene length. All tag-counts were normalized by the total number of bases in the windows and the total read number of the given library to obtain normalized tag density (Wang et al., 2008). (D) The percentage of genes associated with H3K4me3 alone, both H3K4me3 and H3K27me3, H3K27me3 alone, or not associated with H3K4me3 or H3K27me3 for each T cell subsets.
Figure 3. Epigenetic modifications and gene expression profiles of signature genes characterizing T cell subsets
H3K4me3 and H3K27me3 modifications as well as gene expression were first normalized separately using GeneSpring software. Hierarchical clusters of signature genes for Th1, Th2, Th17, iTreg, and nTreg cells are shown. The color coding depicts the normalized value for each gene based on a scale of 0-6 with 1 as the median. The cell types for the expression and modification patterns are indicated below the panels. Each cluster is characteristic of one cell type, which is indicated on the left of the panel. The shared genes between iTreg and nTreg cells are indicated by asterisks on right.
Figure 4. Lineage specific alternative transcript usage and functional regulatory elements
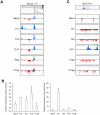
(A) Distribution of H3K4me3 and H3K27me3 islands along the Runx3 genomic region (Chr4, 134389000-134451000) in six T cell subsets is shown. All figures with views of H3K4me3 and H3K27me3 distribution are labeled such that the arrow represents the direction of gene transcription. Gene structure is downloaded from UCSC Genome Browser and only tags on islands are shown. The islands labeled in blue represent H3K4me3. The islands labeled in red represent H3K27me3. Scales are kept constant among cell types. (B) Real-time PCR was performed on RNAs from naïve, Th1, Th2, Th17, and iTreg cells using primer sets that recognize both the long and short forms (left panel) or only the long form (right panel) of Runx3 transcripts. (C) Distribution of H3K4me3 and H3K27me islands along the _I_ntergenic _R_egion upstream of Il17f (IR-Il17f) (Chr1, 20,755,000-20,780,000) in six T cell subsets is shown. The intergenic H3K4me3 island is highlighted by a red circle.
Figure 5. H3K4me3 and H3K27me3 modifications of signature cytokine and transcription factor genes
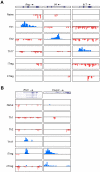
(A) H3K4me3 (blue) and H3K27me3 (red) modifications on Ifng (Chr10, 117842000-117850000), Il4 (Chr11, 53455000-53465000), and Il17 (Chr1, 20714000-20722000) loci. (B) H3K4me3 and H3K27me3 modifications on Rorc (Chr3, 94443000-94493000) and Foxp3 (ChrX, 6735380-6753000) loci.
Figure 6. H3K4me3 and H3K27me3 modifications of key transcription factor genes
(A) H3K4me3 (blue) and H3K27me3 (red) modifications on Tbx21 (Chr11, 96894000-96939000) and Gata3 (Chr2, 9770000-9830000) loci. (B,C) Freshly isolated nTreg cells were repolarized under neutral condition (ThN) and Th1 conditions for 72 hours. Proportions of Foxp3- and IFN-γ-producing cells (B) and T-bet (C) were determined by intracellular staining and flow cytometry. The data are representative of two independent experiments.
Comment in
- Programming perpetual T helper cell plasticity.
Rowell E, Wilson CB. Rowell E, et al. Immunity. 2009 Jan 16;30(1):7-9. doi: 10.1016/j.immuni.2008.12.012. Immunity. 2009. PMID: 19144312
Similar articles
- An epigenetic silencing pathway controlling T helper 2 cell lineage commitment.
Allan RS, Zueva E, Cammas F, Schreiber HA, Masson V, Belz GT, Roche D, Maison C, Quivy JP, Almouzni G, Amigorena S. Allan RS, et al. Nature. 2012 Jul 12;487(7406):249-53. doi: 10.1038/nature11173. Nature. 2012. PMID: 22763435 - Regulation of CD4 T-cell differentiation and inflammation by repressive histone methylation.
Antignano F, Zaph C. Antignano F, et al. Immunol Cell Biol. 2015 Mar;93(3):245-52. doi: 10.1038/icb.2014.115. Epub 2015 Jan 13. Immunol Cell Biol. 2015. PMID: 25582341 Review. - The histone H3 lysine-27 demethylase Jmjd3 plays a critical role in specific regulation of Th17 cell differentiation.
Liu Z, Cao W, Xu L, Chen X, Zhan Y, Yang Q, Liu S, Chen P, Jiang Y, Sun X, Tao Y, Hu Y, Li C, Wang Q, Wang Y, Chen CD, Shi Y, Zhang X. Liu Z, et al. J Mol Cell Biol. 2015 Dec;7(6):505-16. doi: 10.1093/jmcb/mjv022. Epub 2015 Apr 3. J Mol Cell Biol. 2015. PMID: 25840993 - Global mapping of H3K4me1 and H3K4me3 reveals the chromatin state-based cell type-specific gene regulation in human Treg cells.
Tian Y, Jia Z, Wang J, Huang Z, Tang J, Zheng Y, Tang Y, Wang Q, Tian Z, Yang D, Zhang Y, Fu X, Song J, Liu S, van Velkinburgh JC, Wu Y, Ni B. Tian Y, et al. PLoS One. 2011;6(11):e27770. doi: 10.1371/journal.pone.0027770. Epub 2011 Nov 23. PLoS One. 2011. PMID: 22132139 Free PMC article. - Plasticity of CD4+ T cell lineage differentiation.
Zhou L, Chong MM, Littman DR. Zhou L, et al. Immunity. 2009 May;30(5):646-55. doi: 10.1016/j.immuni.2009.05.001. Immunity. 2009. PMID: 19464987 Review.
Cited by
- Epigenetic regulation in epithelial cells and innate lymphocyte responses to S. Typhi infection: insights into IFN-γ production and intestinal immunity.
Salerno-Goncalves R, Chen H, Bafford AC, Sztein MB. Salerno-Goncalves R, et al. Front Immunol. 2024 Sep 20;15:1448717. doi: 10.3389/fimmu.2024.1448717. eCollection 2024. Front Immunol. 2024. PMID: 39372404 Free PMC article. - Transcriptional re-programming of liver-resident iNKT cells into T-regulatory type-1-like liver iNKT cells involves extensive gene de-methylation.
Montaño J, Garnica J, Yamanouchi J, Moro J, Solé P, Mondal D, Serra P, Yang Y, Santamaria P. Montaño J, et al. Front Immunol. 2024 Sep 9;15:1454314. doi: 10.3389/fimmu.2024.1454314. eCollection 2024. Front Immunol. 2024. PMID: 39315110 Free PMC article. - Bivalent chromatin accommodates survivin and BRG1/SWI complex to activate DNA damage response in CD4+ cells.
Chandrasekaran V, Andersson KME, Erlandsson M, Li S, Olsson TN, Garcia-Bonete MJ, Malmhäll-Bah E, Johansson P, Katona G, Bokarewa MI. Chandrasekaran V, et al. Cell Commun Signal. 2024 Sep 11;22(1):440. doi: 10.1186/s12964-024-01814-4. Cell Commun Signal. 2024. PMID: 39261837 Free PMC article. - Epigenetic regulation of human FOXP3+ Tregs: from homeostasis maintenance to pathogen defense.
Yue Y, Ren Y, Lu C, Li P, Zhang G. Yue Y, et al. Front Immunol. 2024 Jul 31;15:1444533. doi: 10.3389/fimmu.2024.1444533. eCollection 2024. Front Immunol. 2024. PMID: 39144146 Free PMC article. Review. - Case report: Macrophage activation syndrome in a patient with Kabuki syndrome.
Zhang J, Kang Y, Xia Z, Chong Y, Long X, Shen M. Zhang J, et al. Front Immunol. 2024 Jul 30;15:1412084. doi: 10.3389/fimmu.2024.1412084. eCollection 2024. Front Immunol. 2024. PMID: 39139573 Free PMC article.
References
- Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat Immunol. 2002;3:549–557. - PubMed
- Akimzhanov AM, Yang XO, Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J Biol Chem. 2007;282:5969–5972. - PubMed
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. - PubMed
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. - PubMed
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials