The CREB/CREM transcription factors negatively regulate early synaptogenesis and spontaneous network activity - PubMed (original) (raw)
The CREB/CREM transcription factors negatively regulate early synaptogenesis and spontaneous network activity
Fernando Aguado et al. J Neurosci. 2009.
Abstract
The family of CREB (cAMP response element-binding protein) transcription factors are involved in a variety of biological processes including the development and plasticity of the nervous system. In the maturing and adult brain, CREB genes are required for activity-dependent processes, including synaptogenesis, refinement of connections and long-term potentiation. Here, we use CREB1(Nescre)CREM(-/-) (cAMP-responsive element modulator) mutants to investigate the role of these genes in stimulus-independent patterns of neural activity at early stages. We show that lack of CREB/CREM genes specifically in neural tissue leads to increased synaptogenesis and to a dramatic increase in the levels of spontaneous network activity at embryonic stages. Thus, the functions of CREB/CREM genes in neural activity differ in distinct periods of neural development.
Figures
Figure 1.
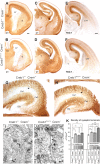
CREB/CREM mutant mice display normal fiber cytoarchitectonics but increased synaptogenesis. A–F, Coronal sections of E18 forebrains immunoreacted for L1 and TAG1 showing that the trajectories of the main fiber tracts of the forebrain are unaltered in CREB1NescreCREM−/− embryos. CC, Corpus callosum; CTT, corticothalamic tract; F, fimbria; H, hippocampus; S, striatum; T, thalamus. G–H, Hippocampal sections immunoreacted for L1 showing similar patterns of fiber distribution (arrows) in CREB1+/+CREM+/− and CREB1NescreCREM−/− embryos. DG, Dentate gyrus; CA1–CA3, hippocampal subfields. I, J, Electron micrographs showing axon terminals displaying synaptic contacts (arrows) in the stratum radiatum of CREB1+/+CREM+/− and CREB1NescreCREM−/− hippocampi. K, Density of synaptic terminals in the stratum radiatum and stratum lacunosum moleculare in the genotypes showing increased number of contacts in CREB1+/+CREM−/−, CREB1NescreCREM+/−, and CREB1NescreCREM−/− embryos, compared with CREB1+/+CREM+/− brains. *p < 0.05, **p < 0.01). Scale bars: A–H, 200 μm; I, J, 0.5 μm.
Figure 2.
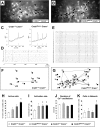
Spontaneous correlated network activity is enhanced in the hippocampus of E18 CREB1NescreCREM+/− and CREB1NescreCREM−/− embryos. A–G, Examples of network activity in the hippocampal CA1 region of CREB1+/+CREM+/− and CREB1NescreCREM−/− embryos. A, B, Fura-2-loaded hippocampal slices showing active neurons (squares). C, Representative plots of Δ_F_/F over time illustrating spontaneous [Ca+2]i changes in a hippocampal neuron. The initiation of [Ca+2]i transients is indicted by bars on the _x_-axis. D, E, Raster plots of all active cells shown in A and B in a CREB1+/+CREM+/− (D) and CREB1NescreCREM−/− (E) slice. Horizontal lines represent the activity profile over 800 s of each active neuron, and vertical thick lines mark the onset of [Ca+2]i oscillations. The long vertical lines crossing the horizontal lines in D and E mark the times where the onset of the oscillations is synchronous across multiple cells. F, G, Correlation maps illustrating a significant (p < 0.01) spatiotemporal coactivation among all cell pairs shown in A–D and B–E assayed with χ2 contingency tables. Each pair of synchronous cells is connected by lines. The thickness of the lines is proportional to the degree of significance. The p value reflects the overall level of coactivation obtained by Monte Carlo simulation and represents the probability that the coactivations present in this field are caused by chance. The complexity of spatiotemporal correlations is enhanced in CREB1NescreCREM−/− embryos, as denoted by the increased number of cells involved in correlated networks. H–K, Histograms showing significant increases in the percentage of active neurons (H), the activation rates Ca2+ transients/cell/104 s (I), and the number of active cells involved in correlated networks in CREB1NescreCREM−/− hippocampal slices (K), whereas durations of Ca2+ oscillation do not differ (J). *p < 0.01. Scale bar, 50 μm.
Figure 3.
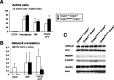
A, Histograms showing the percentage of active neurons (with respect to controls) in the E18 CA1 hippocampal region, after incubations with EGTA, thapsigargin, BMI, and CNQX/APV. White bars represent control CREB1+/+CREM+/− slices and black bars represent CREB1nescreCREM−/− double-mutant embryos. B, Average of Monte Carlo p values representing the probability that the overall degree of synchronous correlation present in the network is caused by chance. CREB1NescreCREM−/− hippocampal slices exhibit significant values in basal conditions, whereas neurotransmitter receptor antagonists decorrelate spontaneous activity. C, Western blots showing protein expression levels of neurotransmitter receptors (GABAAα1 subunit, glutamate receptor NMDA receptor subunit NR1 and GluR2/3) in brain lysates of E18 embryos. Densitometric analyses revealed no significant differences among groups in neurotransmitter receptor protein expression levels (p > 0.05).
Comment in
- A Janus-like role of CREB protein: enhancement of synaptic property in mature neurons and suppression of synaptogenesis and reduced network synchrony in early development.
Nonaka M. Nonaka M. J Neurosci. 2009 May 20;29(20):6389-91. doi: 10.1523/JNEUROSCI.1309-09.2009. J Neurosci. 2009. PMID: 19458209 Free PMC article. Review. No abstract available.
Similar articles
- Regulation of neural migration by the CREB/CREM transcription factors and altered Dab1 levels in CREB/CREM mutants.
Díaz-Ruiz C, Parlato R, Aguado F, Ureña JM, Burgaya F, Martínez A, Carmona MA, Kreiner G, Bleckmann S, Del Río JA, Schütz G, Soriano E. Díaz-Ruiz C, et al. Mol Cell Neurosci. 2008 Dec;39(4):519-28. doi: 10.1016/j.mcn.2008.07.019. Epub 2008 Aug 5. Mol Cell Neurosci. 2008. PMID: 18786638 - Survival of DA neurons is independent of CREM upregulation in absence of CREB.
Parlato R, Rieker C, Turiault M, Tronche F, Schütz G. Parlato R, et al. Genesis. 2006 Oct;44(10):454-64. doi: 10.1002/dvg.20236. Genesis. 2006. PMID: 16981198 - Effects of the cell type-specific ablation of the cAMP-responsive transcription factor in noradrenergic neurons on locus coeruleus firing and withdrawal behavior after chronic exposure to morphine.
Parlato R, Cruz H, Otto C, Murtra P, Parkitna JR, Martin M, Bura SA, Begus-Nahrmann Y, von Bohlen und Halbach O, Maldonado R, Schütz G, Lüscher C. Parlato R, et al. J Neurochem. 2010 Nov;115(3):563-73. doi: 10.1111/j.1471-4159.2010.06709.x. Epub 2010 Aug 19. J Neurochem. 2010. PMID: 20367754 - The expanding family of CREB/CREM transcription factors that are involved with spermatogenesis.
Don J, Stelzer G. Don J, et al. Mol Cell Endocrinol. 2002 Feb 22;187(1-2):115-24. doi: 10.1016/s0303-7207(01)00696-7. Mol Cell Endocrinol. 2002. PMID: 11988318 Review. - Coupling cAMP signaling to transcription in the liver: pivotal role of CREB and CREM.
Servillo G, Della Fazia MA, Sassone-Corsi P. Servillo G, et al. Exp Cell Res. 2002 May 1;275(2):143-54. doi: 10.1006/excr.2002.5491. Exp Cell Res. 2002. PMID: 11969286 Review.
Cited by
- Inducible forebrain-specific ablation of the transcription factor Creb during adulthood induces anxiety but no spatial/contextual learning deficits.
Vogt MA, Inta D, Luoni A, Elkin H, Pfeiffer N, Riva MA, Gass P. Vogt MA, et al. Front Behav Neurosci. 2014 Nov 27;8:407. doi: 10.3389/fnbeh.2014.00407. eCollection 2014. Front Behav Neurosci. 2014. PMID: 25505876 Free PMC article. - Molecular mechanisms of synaptic remodeling in alcoholism.
Kyzar EJ, Pandey SC. Kyzar EJ, et al. Neurosci Lett. 2015 Aug 5;601:11-9. doi: 10.1016/j.neulet.2015.01.051. Epub 2015 Jan 23. Neurosci Lett. 2015. PMID: 25623036 Free PMC article. Review. - A Janus-like role of CREB protein: enhancement of synaptic property in mature neurons and suppression of synaptogenesis and reduced network synchrony in early development.
Nonaka M. Nonaka M. J Neurosci. 2009 May 20;29(20):6389-91. doi: 10.1523/JNEUROSCI.1309-09.2009. J Neurosci. 2009. PMID: 19458209 Free PMC article. Review. No abstract available. - Biomechanical Forces Regulate Gene Transcription During Stretch-Mediated Growth of Mammalian Neurons.
Loverde JR, Tolentino RE, Soteropoulos P, Pfister BJ. Loverde JR, et al. Front Neurosci. 2020 Dec 8;14:600136. doi: 10.3389/fnins.2020.600136. eCollection 2020. Front Neurosci. 2020. PMID: 33408609 Free PMC article. - Toxicogenomic profiling in maternal and fetal rodent brains following gestational exposure to chlorpyrifos.
Moreira EG, Yu X, Robinson JF, Griffith W, Hong SW, Beyer RP, Bammler TK, Faustman EM. Moreira EG, et al. Toxicol Appl Pharmacol. 2010 Jun 15;245(3):310-25. doi: 10.1016/j.taap.2010.03.015. Epub 2010 Mar 27. Toxicol Appl Pharmacol. 2010. PMID: 20350560 Free PMC article.
References
- Aguado F, Carmona MA, Pozas E, Aguiló A, Martínez-Guijarro FJ, Alcantara S, Borrell V, Yuste R, Ibañez CF, Soriano E. BDNF regulates spontaneous correlated activity at early developmental stages by increasing synaptogenesis and expression of the K+/Cl- co-transporter KCC2. Development. 2003;130:1267–1280. - PubMed
- Barco A, Alarcon JM, Kandel ER. Expression of constitutively active CREB protein facilitates the late phase of long-term potentiation by enhancing synaptic capture. Cell. 2002;108:689–703. - PubMed
- Ben-Ari Y. Developing networks play a similar melody. Trends Neurosci. 2001;24:353–360. - PubMed
- Demas J, Sagdullaev BT, Green E, Jaubert-Miazza L, McCall MA, Gregg RG, Wong RO, Guido W. Failure to maintain eye-specific segregation in nob, a mutant with abnormally patterned retinal activity. Neuron. 2006;50:247–259. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases