Two Piwi proteins, Xiwi and Xili, are expressed in the Xenopus female germline - PubMed (original) (raw)
Two Piwi proteins, Xiwi and Xili, are expressed in the Xenopus female germline
Anna Wilczynska et al. RNA. 2009 Feb.
Abstract
The Argonaute superfamily is a large family of RNA-binding proteins involved in gene regulation mediated by small noncoding RNA and characterized by the presence of PAZ and PIWI domains. The family consists of two branches, the Ago and the Piwi clade. Piwi proteins bind to 21-30-nucleotide-long Piwi-interacting RNAs (piRNAs), which map primarily to transposons and repeated sequence elements. Piwi/piRNAs are important regulators of gametogenesis and have been proposed to play roles in transposon silencing, DNA methylation, transcriptional silencing, and/or post-transcriptional control of translation and RNA stability. Most reports to date have concentrated on the Piwi family members in the male germline. We have identified four Piwi proteins in Xenopus and demonstrate that two, namely, Xiwi1b and Xili, are expressed in the oocyte and early embryo. Xiwi1 and Xili are predominantly found in small, separate complexes, and we do not detect significant interaction of Piwi proteins with the cap-binding complex. Putative nuclear localization and export signals were identified in Xiwi1 and Xili, supporting our observation that Xiwi1, but not Xili, is a nucleo-cytoplasmic protein. Furthermore, by immunoprecipitation of small RNAs, we establish Xiwi1 as a bona fide Piwi protein. These results suggest that the Piwi/piRNA pathway is active in translationally repressed oocytes. This is a significant finding as the Xenopus model provides an excellent tool to study post-transcriptional mechanisms.
Figures
FIGURE 1.
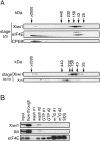
Identification of Xenopus Piwi proteins. (A) Phylogenetic tree of Piwi family proteins. Unrooted phylogenetic tree based on ClustalW alignment of Piwi domains as predicted by the Prosite database (Hulo et al. 2008). The sequences used are PRG-1, C. elegans NM_059720; PRG-2, C. elegans NM_068593; Piwi, D. melanogaster NM_057527; Aubergine, D. melanogaster NM_057386; DmAgo3, D. melanogaster EF211827; Seawi, S. purpuratus NM_214600; Ziwi, D. rerio NM_183338; Zili, D. rerio EF186090; Miwi, M. musculus NM_021311; Miwi2, M. musculus NM_177905; Mili, M. musculus NM_021308; Hiwi, H. sapiens NM_004764; Hiwi2, H. sapiens NM_152431; Hili, H. sapiens NM_018068; Hiwi3, H. sapiens NM_001008496. (B) ClustalW alignment of four putative Xenopus tropicalis Xiwi1a, Xiwi1b, Xili, and Xiwi2 protein sequences. The Xiwi1a sequence was derived from IMAGE clone 7689388. The Xiwi1b sequence was assembled from scaffolds Xt7.1-CAAN9738.3.5 and Xt7.1-XZG3113.5 of the Gurdon Institute X. tropicalis full-length database. Xili was derived from IMAGE clone 7676520, and Xiwi2 was assembled from scaffolds Xt7.1-CAAN6696.5, Xt7.1-CAAN4060.3, and Ensembl Gene prediction ENSXETP00000014018. (Highly related, albeit partial, X. laevis EST sequences were also retrieved from databases.) Sequences highlighted with the solid gray box denote peptides used for generating specific antibodies against Xiwi1 and Xili, respectively. (Dashed black box) PAZ and (solid black box) PIWI domains. (C) Antibodies raised against Xiwi1 and Xili are specific to the proteins and do not cross react. Polyclonal antibodies were tested on a Western blot containing one oocyte equivalent of stage VI Xenopus oocyte lysate, 4.5 ng of recombinant His-tagged Xili protein, and 1.5 ng of recombinant His-tagged Xiwi1. (*) Nonspecific bands detected by the antibodies. Proteins were resolved on a 15% polyacrylamide SDS gel.
FIGURE 2.
Expression of Xiwi1 and Xili throughout Xenopus oogenesis and embryogenesis. Two oocyte equivalents for each stage of oogenesis, two egg equivalents, two embryo equivalents for each stage of embryogenesis, and 20 μg of testis lysate protein (equivalent of one stage VI oocyte) were resolved on 10% SDS-PAGE. The stages of embryogenesis correspond to stage 9, midblastula; stage 12.5, gastrula; stage 20, neural fold closure; stage 26, tail bud stage; stage 42, tadpole-like stage. Stage I may be underrepresented here because of the technical difficulties of accurately estimating cell number with these small cells. An alternative image of Xiwi1 protein in testis is presented for better visualization of the protein doublet. Note that Xili appears as a doublet in oocytes, for unknown reasons, due to better resolution of high-molecular-weight proteins compared to Figure 1C.
FIGURE 3.
(A) Xiwi1 and Xili are predominantly present in small complexes. Lysate prepared from (upper panel) stage I/II and (lower panel) stage III/IV oocytes was gel-filtered as described previously, on separate columns (Minshall et al. 2007). Alternate fractions were resolved by 15% SDS-PAGE and analyzed by Western blot with the indicated antibodies. For stage I/II fractions, Xiwi1 was detected using the anti-Miwi antibody (Deng and Lin 2002). For stage III/IV fractions, the Western blot was probed with the anti-Xiwi1 and anti-Xili antibodies described above. The molecular weight standards used were chymotrypsin A (25 kDa), ovalbumin (43 kDa), aldolase (158 kDa), catalase (232 kDa), and ferritin (440 kDa). (B) Xiwi1 and Xili do not interact with the cap-binding complex. m7GTP-Sepharose chromatography was performed using stage III/IV oocyte lysate. Following binding, the beads were washed and then eluted with GTP- and m7GpppG-containing buffer and finally with SDS buffer. Input represents ∼0.25% of the input lysate; the flow-through and wash fractions represent 1% of each fraction; the GTP and m7G fractions represent 12% of each fraction; and SDS represents 50% of the SDS fraction.
FIGURE 4.
Xiwi1 localization is partially nuclear, while Xili is exclusively present in the cytoplasm. (A) Equivalents of (T) one total stage VI oocyte, (C) one cytoplasm, (N) one nucleus, and (Nx2) two nuclei were resolved on 10% SDS-PAGE and analyzed by Western blot. PARN was used to assess the content of the nuclear fraction, and CPEB was used as a marker of the cytoplasm. Xili appears as a doublet due to better resolution of high-molecular-weight proteins compared to Figure 1C. PARN was used to assess the content of the nuclear fraction, and CPEB was used as a marker of the cytoplasm. (B) Predicted (NES) nuclear export signals were identified in the coding sequence of both Xiwi1 (616MGGELWSVEI625) and Xili (258LPVKLAQTVEL268, 304MRRVFKILDLKL313, 490MKDLTQQIHL499, 701LGGELWGVDI710), according to the consensus (L/M)-X2-3-(L/F)- X2-3-(L/I/V)-X-(L/I) (Kutay and Guttinger 2005). One putative nuclear localization signal (NLS) was identified in the coding sequence of Xiwi1 (4RARARARGRARG15) using NLSdb (Nair et al. 2003).
FIGURE 5.
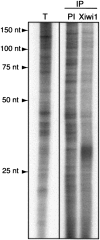
Xiwi1 interacts specifically with RNAs ∼30 nt in length. RNAs immunoprecipitated with an antibody directed against Xiwi1 and the corresponding preimmune serum and total small RNA were extracted and 32P-end-labeled. A specific fraction migrating at ∼30 nt is enriched in anti-Xiwi immunoprecipitates. (IP) Immunoprecipitated samples; (PI) immunoprecipitation with preimmune serum corresponding to the anti-Xiwi serum. Total small RNA was isolated using the mirVana kit (Ambion).
Similar articles
- Xenopus Piwi proteins interact with a broad proportion of the oocyte transcriptome.
Toombs JA, Sytnikova YA, Chirn GW, Ang I, Lau NC, Blower MD. Toombs JA, et al. RNA. 2017 Apr;23(4):504-520. doi: 10.1261/rna.058859.116. Epub 2016 Dec 28. RNA. 2017. PMID: 28031481 Free PMC article. - Systematic and single cell analysis of Xenopus Piwi-interacting RNAs and Xiwi.
Lau NC, Ohsumi T, Borowsky M, Kingston RE, Blower MD. Lau NC, et al. EMBO J. 2009 Oct 7;28(19):2945-58. doi: 10.1038/emboj.2009.237. Epub 2009 Aug 27. EMBO J. 2009. PMID: 19713941 Free PMC article. - The biogenesis and function of PIWI proteins and piRNAs: progress and prospect.
Thomson T, Lin H. Thomson T, et al. Annu Rev Cell Dev Biol. 2009;25:355-76. doi: 10.1146/annurev.cellbio.24.110707.175327. Annu Rev Cell Dev Biol. 2009. PMID: 19575643 Free PMC article. Review. - PIWI Slicing and EXD1 Drive Biogenesis of Nuclear piRNAs from Cytosolic Targets of the Mouse piRNA Pathway.
Yang Z, Chen KM, Pandey RR, Homolka D, Reuter M, Janeiro BK, Sachidanandam R, Fauvarque MO, McCarthy AA, Pillai RS. Yang Z, et al. Mol Cell. 2016 Jan 7;61(1):138-52. doi: 10.1016/j.molcel.2015.11.009. Epub 2015 Dec 6. Mol Cell. 2016. PMID: 26669262 Free PMC article. - Advances in PIWI-piRNA function in female reproduction in mammals.
Lv X, Zhang H, Wu L. Lv X, et al. Acta Biochim Biophys Sin (Shanghai). 2024 Nov 15;57(1):148-156. doi: 10.3724/abbs.2024195. Acta Biochim Biophys Sin (Shanghai). 2024. PMID: 39544003 Free PMC article. Review.
Cited by
- PIWI Expression and Function in Cancer.
Suzuki R, Honda S, Kirino Y. Suzuki R, et al. Front Genet. 2012 Oct 16;3:204. doi: 10.3389/fgene.2012.00204. eCollection 2012. Front Genet. 2012. PMID: 23087701 Free PMC article. - Pinpointing the expression of piRNAs and function of the PIWI protein subfamily during spermatogenesis in the mouse.
Beyret E, Lin H. Beyret E, et al. Dev Biol. 2011 Jul 15;355(2):215-26. doi: 10.1016/j.ydbio.2011.04.021. Epub 2011 Apr 22. Dev Biol. 2011. PMID: 21539824 Free PMC article. - Establishing, maintaining and modifying DNA methylation patterns in plants and animals.
Law JA, Jacobsen SE. Law JA, et al. Nat Rev Genet. 2010 Mar;11(3):204-20. doi: 10.1038/nrg2719. Nat Rev Genet. 2010. PMID: 20142834 Free PMC article. Review. - Expansion of genes encoding piRNA-associated argonaute proteins in the pea aphid: diversification of expression profiles in different plastic morphs.
Lu HL, Tanguy S, Rispe C, Gauthier JP, Walsh T, Gordon K, Edwards O, Tagu D, Chang CC, Jaubert-Possamai S. Lu HL, et al. PLoS One. 2011;6(12):e28051. doi: 10.1371/journal.pone.0028051. Epub 2011 Dec 5. PLoS One. 2011. PMID: 22162754 Free PMC article. - Evolutionary history of the vertebrate Piwi gene family.
Gutierrez J, Platt R, Opazo JC, Ray DA, Hoffmann F, Vandewege M. Gutierrez J, et al. PeerJ. 2021 Nov 5;9:e12451. doi: 10.7717/peerj.12451. eCollection 2021. PeerJ. 2021. PMID: 34760405 Free PMC article.
References
- Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. - PubMed
- Aravin A.A., Hannon G.J., Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007a;318:761–764. - PubMed
- Aravin A.A., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G.J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007b;316:744–747. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous