The human intestinal microbiome: a new frontier of human biology - PubMed (original) (raw)
Review
The human intestinal microbiome: a new frontier of human biology
Masahira Hattori et al. DNA Res. 2009 Feb.
Abstract
To analyze the vast number and variety of microorganisms inhabiting the human intestine, emerging metagenomic technologies are extremely powerful. The intestinal microbes are taxonomically complex and constitute an ecologically dynamic community (microbiota) that has long been believed to possess a strong impact on human physiology. Furthermore, they are heavily involved in the maturation and proliferation of human intestinal cells, helping to maintain their homeostasis and can be causative of various diseases, such as inflammatory bowel disease and obesity. A simplified animal model system has provided the mechanistic basis for the molecular interactions that occur at the interface between such microbes and host intestinal epithelia. Through metagenomic analysis, it is now possible to comprehensively explore the genetic nature of the intestinal microbiome, the mutually interacting system comprising the host cells and the residing microbial community. The human microbiome project was recently launched as an international collaborative research effort to further promote this newly developing field and to pave the way to a new frontier of human biology, which will provide new strategies for the maintenance of human health.
Figures
Figure 1
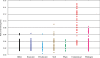
Ratio of gut microbiome-enriched genes (adult) in sequenced genomes of bacteria isolated from various environments. The similarity search was performed with a threshold value of ≤1e−8 for 371 bacteria whose genomic sequences were available from public databases. The 371 bacteria were classified into seven groups according to their origin of isolation; commensals, pathogens, plant-related, soil-born, freshwater-born, seawater-born and others, and shown in red, pink, green, brown, light blue, dark blue and black dots, respectively.
Figure 2
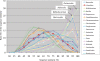
Distribution of sequence similarity of genes identified in the human intestinal microbiome. Only the data for the species that had sufficient numbers of best blastp-hit to known genes were represented. Species names are indicated in the box. Of these, four typical distribution patterns, which peaked at high sequence similarity (≥80% ID), are indicated by name in the figure.
Figure 3
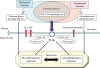
Molecular interactions at the frontline between the host, intestinal commensal bacteria and pathogenic bacteria. Commensal bacteria possess specific functions adaptive to the gut habitat and are beneficial to host cells, including maintenance of intestinal homeostasis, but also include functions such as those of TLRs that signal immune responses to pathogenic infection., Host cells produce various antimicrobial substances such as IgA, IAP and antimicrobial peptides at the frontline to suppress excessive immune response to commensal bacteria, while maintaining responsiveness to pathogens equipped with various virulence factors to evade the host defense system.
Figure 4
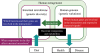
Future direction of human intestinal microbiota research. Human intestinal microbiota function is responsible for both human health and disease in accordance with its own genetic diversity and in association with human genetic variation. The study of human microbes, especially the vastly abundant intestinal microbes, is a new frontier in human biology. Many questions remain to be answered about host–microbe interactions, including: what factors and dietary components shape microbiota diversity, which bacteria and their components interact with host cells, which human genes respond to and how do they react to bacterial signals affecting human physiology.
Similar articles
- [Disease and metagenomics of intestinal microbiomes].
Hattori M. Hattori M. Nihon Rinsho. 2009 Jun;67(6):1214-8. Nihon Rinsho. 2009. PMID: 19507517 Japanese. - Interactions between intestinal microbiota and innate immune system in pediatric inflammatory bowel disease.
Cucchiara S, Stronati L, Aloi M. Cucchiara S, et al. J Clin Gastroenterol. 2012 Oct;46 Suppl:S64-6. doi: 10.1097/MCG.0b013e31826a857f. J Clin Gastroenterol. 2012. PMID: 22955361 Retracted. - Host-microbiota interactions in inflammatory bowel disease.
Caruso R, Lo BC, Núñez G. Caruso R, et al. Nat Rev Immunol. 2020 Jul;20(7):411-426. doi: 10.1038/s41577-019-0268-7. Epub 2020 Jan 31. Nat Rev Immunol. 2020. PMID: 32005980 Review. - [Interaction between humans and intestinal bacteria as a determinant for intestinal health : intestinal microbiome and inflammatory bowel diseases].
Haller D, Hörmannsperger G. Haller D, et al. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2015 Feb;58(2):159-65. doi: 10.1007/s00103-014-2095-0. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2015. PMID: 25566836 Review. German. - Influences of intestinal bacteria in human inflammatory bowel disease.
Vanderploeg R, Panaccione R, Ghosh S, Rioux K. Vanderploeg R, et al. Infect Dis Clin North Am. 2010 Dec;24(4):977-93, ix. doi: 10.1016/j.idc.2010.07.008. Infect Dis Clin North Am. 2010. PMID: 20937461 Review.
Cited by
- Gut Microbiota: A Contributing Factor to Obesity.
Harakeh SM, Khan I, Kumosani T, Barbour E, Almasaudi SB, Bahijri SM, Alfadul SM, Ajabnoor GM, Azhar EI. Harakeh SM, et al. Front Cell Infect Microbiol. 2016 Aug 30;6:95. doi: 10.3389/fcimb.2016.00095. eCollection 2016. Front Cell Infect Microbiol. 2016. PMID: 27625997 Free PMC article. Review. - Modifier-concept of colorectal carcinogenesis: lipidomics as a technical tool in pathway analysis.
Gassler N, Klaus C, Kaemmerer E, Reinartz A. Gassler N, et al. World J Gastroenterol. 2010 Apr 21;16(15):1820-7. doi: 10.3748/wjg.v16.i15.1820. World J Gastroenterol. 2010. PMID: 20397257 Free PMC article. - Characteristics of Gorilla-Specific Lactobacillus Isolated from Captive and Wild Gorillas.
Tsuchida S, Kakooza S, Mbehang Nguema PP, Wampande EM, Ushida K. Tsuchida S, et al. Microorganisms. 2018 Aug 14;6(3):86. doi: 10.3390/microorganisms6030086. Microorganisms. 2018. PMID: 30110987 Free PMC article. - Nodeomics: pathogen detection in vertebrate lymph nodes using meta-transcriptomics.
Wittekindt NE, Padhi A, Schuster SC, Qi J, Zhao F, Tomsho LP, Kasson LR, Packard M, Cross P, Poss M. Wittekindt NE, et al. PLoS One. 2010 Oct 18;5(10):e13432. doi: 10.1371/journal.pone.0013432. PLoS One. 2010. PMID: 20976145 Free PMC article. - Manipulation of the intestinal microbiome in newborn infants.
Cacho N, Neu J. Cacho N, et al. Adv Nutr. 2014 Jan 1;5(1):114-8. doi: 10.3945/an.113.004820. Adv Nutr. 2014. PMID: 24425730 Free PMC article. Review.
References
- Savage D. C. Microbial ecology of the gastrointestinal tract. Ann. Rev. Microbiol. 1977;31:107–133. - PubMed
- Lederberg J. Infectious history. Science. 2000;288:287–293. - PubMed
- Bäckhed F., et al. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. - PubMed
- Ley R. E., et al. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources