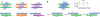The molecular basis of TCR germline bias for MHC is surprisingly simple - PubMed (original) (raw)
Review
The molecular basis of TCR germline bias for MHC is surprisingly simple
K Christopher Garcia et al. Nat Immunol. 2009 Feb.
Abstract
The elusive etiology of germline bias of the T cell receptor (TCR) for major histocompatibility complex (MHC) has been clarified by recent 'proof-of-concept' structural results demonstrating the conservation of specific TCR-MHC interfacial contacts in complexes bearing common variable segments and MHC allotypes. We suggest that each TCR variable-region gene product engages each type of MHC through a 'menu' of structurally coded recognition motifs that have arisen through coevolution. The requirement for MHC-restricted T cell recognition during thymic selection and peripheral surveillance has necessitated the existence of such a coded recognition system. Given these findings, a reconsideration of the TCR-peptide-MHC structural database shows that not only have the answers been there all along but also they were predictable by the first principles of physical chemistry.
Figures
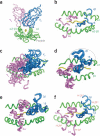
Figure 1
Divergence and convergence of TCR footprints on MHC molecules. (a) Interaction of VαVβ with peptide-MHC, viewed down the MHC groove (Protein Data Bank accession number, 2CKB). (b) ’Footprint‘ view of a showing the stereotyped polarity of the Vα and Vβ CDR loops on pMHC. (c) Convergent footprint polarity but diverse CDR loop positions in nine different TCR-pMHC complexes (Protein Data Bank accession numbers, 1AO7, 1FO0, 1J8H, 1KJ2, 1ZG1, 2NX5, 1MI5, 1OGA and 1U3H). (d) Close superpositions (in circle) of the contacts of Vβ8 CDR1 and CDR2 with the I-A MHC α1 helix in six different TCR-pMHC complexes (Protein Data Bank accession numbers, 1U3H, 2Z31, 2PXY, 1D9K, 3C60 and 3C61). (e) Retention of similar germline-mediated contacts by the BM3.3 TCR with H-2Kb in three different peptide complexes (Protein Data Bank accession numbers, 1FO0, 2OL3 and 1NAM). (f) Use of alternative ‘codons’ for interaction of the 2C TCR Vα and Vβ with H-2Kb versus H-2Ld (Protein Data Bank accession numbers, 2CKB and 2OI9). H-2Kb and H-2Ld (in red) adjacent to the respective loops indicate the positions of CDR2α and CDR2β in the structures.
Figure 2
The ‘codon hypothesis’ for germline TCR-MHC interactions. (a) each V-gene product (where ‘Vx’ is Vα or Vβ) interacts with diverse MHC surface residues on the tops of the helices of different MHC molecules (Y, X and Z) by distinct yet specific mechanisms. That is, each CDR1 and/or CDR2 engages different MHC surfaces in diverse ways: the pairwise interactions that form each codon need not be shared by different MHC molecules. The CDR-MHC interface is presented as ‘teeth’ on the respective interacting surfaces. The same ‘teeth’ in a particular Vα or Vβ CDR1-CDR2 engage different MHC surface structures (opposing ‘teeth’) in each complex in unique, highly specific ways. (b) An individual TCR germline V segment can engage one MHC molecule using several distinct codons (A, B and C) that are influenced by the interactions of CDR3 with the MHC-bound peptide. This is presented here as different docking geometries (‘footprints’) on the MHC that are mediated by the interaction of common residues on the TCR CDR1-CDR2 with different ‘registers’ of the MHC helix in each peptide complex. Inset, free energy diagram indicating that each footprint represents a low-energy binding solution (‘click’) rather than an energetic continuum.
Similar articles
- Thymic Origins of T Cell Receptor Alloreactivity.
Brzostek J, Gascoigne NRJ. Brzostek J, et al. Transplantation. 2017 Jul;101(7):1535-1541. doi: 10.1097/TP.0000000000001654. Transplantation. 2017. PMID: 28114168 Review. - Structural and dynamic control of T-cell receptor specificity, cross-reactivity, and binding mechanism.
Baker BM, Scott DR, Blevins SJ, Hawse WF. Baker BM, et al. Immunol Rev. 2012 Nov;250(1):10-31. doi: 10.1111/j.1600-065X.2012.01165.x. Immunol Rev. 2012. PMID: 23046120 Review. - Reconciling views on T cell receptor germline bias for MHC.
Garcia KC. Garcia KC. Trends Immunol. 2012 Sep;33(9):429-36. doi: 10.1016/j.it.2012.05.005. Epub 2012 Jul 6. Trends Immunol. 2012. PMID: 22771140 Free PMC article. - Molecular constraints on CDR3 for thymic selection of MHC-restricted TCRs from a random pre-selection repertoire.
Lu J, Van Laethem F, Bhattacharya A, Craveiro M, Saba I, Chu J, Love NC, Tikhonova A, Radaev S, Sun X, Ko A, Arnon T, Shifrut E, Friedman N, Weng NP, Singer A, Sun PD. Lu J, et al. Nat Commun. 2019 Mar 4;10(1):1019. doi: 10.1038/s41467-019-08906-7. Nat Commun. 2019. PMID: 30833553 Free PMC article. - Regulatory T cells and co-evolution of allele-specific MHC recognition by the TCR.
Steele EJ, Lindley RA. Steele EJ, et al. Scand J Immunol. 2020 Mar;91(3):e12853. doi: 10.1111/sji.12853. Epub 2019 Dec 17. Scand J Immunol. 2020. PMID: 31793005 Free PMC article. Review.
Cited by
- Evolving understanding of autoimmune mechanisms and new therapeutic strategies of autoimmune disorders.
Song Y, Li J, Wu Y. Song Y, et al. Signal Transduct Target Ther. 2024 Oct 4;9(1):263. doi: 10.1038/s41392-024-01952-8. Signal Transduct Target Ther. 2024. PMID: 39362875 Free PMC article. Review. - TCR-H: explainable machine learning prediction of T-cell receptor epitope binding on unseen datasets.
T RR, Demerdash ONA, Smith JC. T RR, et al. Front Immunol. 2024 Aug 16;15:1426173. doi: 10.3389/fimmu.2024.1426173. eCollection 2024. Front Immunol. 2024. PMID: 39221256 Free PMC article. - Selection, engineering, and in vivo testing of a human leukocyte antigen-independent T-cell receptor recognizing human mesothelin.
Hiscox MJ, Wasmuth A, Williams CL, Foot JN, Wiedermann GE, Fadda V, Boiani S, Cornforth TV, Wikiert KA, Bruton S, Cartwright N, Anderson VE, Barnes CS, Vieira JV, Birch-Machin I, Gerry AB, Miller K, Pumphrey NJ. Hiscox MJ, et al. PLoS One. 2024 Apr 4;19(4):e0301175. doi: 10.1371/journal.pone.0301175. eCollection 2024. PLoS One. 2024. PMID: 38574067 Free PMC article. - Cross-reactive CD8+ T cell responses to tumor-associated antigens (TAAs) and homologous microbiota-derived antigens (MoAs).
Cavalluzzo B, Viuff MC, Tvingsholm SA, Ragone C, Manolio C, Mauriello A, Buonaguro FM, Tornesello ML, Izzo F, Morabito A, Hadrup SR, Tagliamonte M, Buonaguro L. Cavalluzzo B, et al. J Exp Clin Cancer Res. 2024 Mar 20;43(1):87. doi: 10.1186/s13046-024-03004-z. J Exp Clin Cancer Res. 2024. PMID: 38509571 Free PMC article. - Conserved biophysical compatibility among the highly variable germline-encoded regions shapes TCR-MHC interactions.
Boughter CT, Meier-Schellersheim M. Boughter CT, et al. Elife. 2023 Oct 20;12:e90681. doi: 10.7554/eLife.90681. Elife. 2023. PMID: 37861280 Free PMC article.
References
- Rudolph MG, stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 2006;24:419–466. - PubMed
- Buslepp J, Wang H, Biddison WE, Appella E, Collins EJ. A correlation between TCR Vα docking on MHC and CD8 dependence: implications for T cell selection. Immunity. 2003;19:595–606. - PubMed
- Collins EJ, Riddle DS. TCR-MHC docking orientation: natural selection, or thymic selection? Immunol. Res. 2008;41:267–294. - PubMed
- Turner SJ, Doherty PC, McCluskey J, Rossjohn J. Structural determinants of T-cell receptor bias in immunity. Nat. Rev. Immunol. 2006;6:883–894. - PubMed
- Tynan FE, et al. T cell receptor recognition of a ‘super-bulged’ major histocompatibility complex class I-bound peptide. Nat. Immunol. 2005;6:1114–1122. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials