Decline in genomic DNA methylation through aging in a cohort of elderly subjects - PubMed (original) (raw)
Decline in genomic DNA methylation through aging in a cohort of elderly subjects
Valentina Bollati et al. Mech Ageing Dev. 2009 Apr.
Abstract
Loss of genomic DNA methylation has been found in a variety of common human age-related diseases. Whether DNA methylation decreases over time as individuals age is unresolved. We measured DNA methylation in 1097 blood DNA samples from 718 elderly subjects between 55 and 92 years of age (1-3 samples/subjects), who have been repeatedly evaluated over an 8-year time span in the Boston area Normative Aging Study. DNA methylation was measured using quantitative PCR-Pyrosequencing analysis in Alu and LINE-1 repetitive elements, heavily methylated sequences with high representation throughout the human genome. Age at the visit was negatively associated with Alu element methylation (beta=-0.12 %5mC/year, p=0.0005). A weaker association was observed with LINE-1 elements (beta=-0.06 %5mC/year, p=0.049). We observed a significant decrease in average Alu methylation over time, with a -0.2 %5mC change (p=0.012) compared to blood samples collected up to 8 years earlier. The longitudinal decline in Alu methylation was linear and highly correlated with time since the first measurement (beta=-0.089 %5mC/year, p<0.0001). In contrast, average LINE-1 methylation did not vary over time [p=0.51]. Our results demonstrate a progressive loss of DNA methylation in repetitive elements dispersed throughout the genome.
Figures
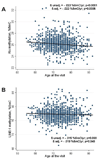
Figure 1
Association between Age and Methylation in Alu (A) and LINE-1 (B) repetitive elements. The graphs show the decrease of methylation (change in % 5Methyl-cytosine [%5mC]/year) in relation to the subject’s age at time of visit. Regression lines and data points represent the unadjusted values. Adjusted and unadjusted Regression coefficients (β) and p-values shown are obtained from unadjusted models and from models adjusted for body mass index, systolic and diastolic blood pressure, fasting blood glucose, percent lymphocytes and neutrophils in blood count, statin use, season of the visit.
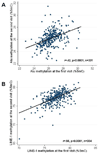
Figure 2
Correlation between the first and the second measurement of Alu (A) and LINE-1 (B) repetitive element methylation taken 2–8 years apart on the same subjects.
Similar articles
- Differential repetitive DNA methylation in multiple myeloma molecular subgroups.
Bollati V, Fabris S, Pegoraro V, Ronchetti D, Mosca L, Deliliers GL, Motta V, Bertazzi PA, Baccarelli A, Neri A. Bollati V, et al. Carcinogenesis. 2009 Aug;30(8):1330-5. doi: 10.1093/carcin/bgp149. Epub 2009 Jun 16. Carcinogenesis. 2009. PMID: 19531770 - Rapid DNA methylation changes after exposure to traffic particles.
Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, Zanobetti A, Sparrow D, Vokonas PS, Schwartz J. Baccarelli A, et al. Am J Respir Crit Care Med. 2009 Apr 1;179(7):572-8. doi: 10.1164/rccm.200807-1097OC. Epub 2009 Jan 8. Am J Respir Crit Care Med. 2009. PMID: 19136372 Free PMC article. - Prolonged exposure to particulate pollution, genes associated with glutathione pathways, and DNA methylation in a cohort of older men.
Madrigano J, Baccarelli A, Mittleman MA, Wright RO, Sparrow D, Vokonas PS, Tarantini L, Schwartz J. Madrigano J, et al. Environ Health Perspect. 2011 Jul;119(7):977-82. doi: 10.1289/ehp.1002773. Epub 2011 Mar 8. Environ Health Perspect. 2011. PMID: 21385671 Free PMC article. - Association between blood pressure and DNA methylation of retrotransposons and pro-inflammatory genes.
Alexeeff SE, Baccarelli AA, Halonen J, Coull BA, Wright RO, Tarantini L, Bollati V, Sparrow D, Vokonas P, Schwartz J. Alexeeff SE, et al. Int J Epidemiol. 2013 Feb;42(1):270-80. doi: 10.1093/ije/dys220. Int J Epidemiol. 2013. PMID: 23508416 Free PMC article. - The epigenetic alterations of endogenous retroelements in aging.
Cardelli M. Cardelli M. Mech Ageing Dev. 2018 Sep;174:30-46. doi: 10.1016/j.mad.2018.02.002. Epub 2018 Feb 16. Mech Ageing Dev. 2018. PMID: 29458070 Review.
Cited by
- Global DNA methylation levels in white blood cells as a biomarker for hepatocellular carcinoma risk: a nested case-control study.
Wu HC, Wang Q, Yang HI, Tsai WY, Chen CJ, Santella RM. Wu HC, et al. Carcinogenesis. 2012 Jul;33(7):1340-5. doi: 10.1093/carcin/bgs160. Epub 2012 May 12. Carcinogenesis. 2012. PMID: 22581841 Free PMC article. - DNA methylation correlates of chronological age in diverse human tissue types.
Jain N, Li JL, Tong L, Jasmine F, Kibriya MG, Demanelis K, Oliva M, Chen LS, Pierce BL. Jain N, et al. Epigenetics Chromatin. 2024 Aug 8;17(1):25. doi: 10.1186/s13072-024-00546-6. Epigenetics Chromatin. 2024. PMID: 39118140 Free PMC article. - Global and gene specific DNA methylation changes during zebrafish development.
Fang X, Corrales J, Thornton C, Scheffler BE, Willett KL. Fang X, et al. Comp Biochem Physiol B Biochem Mol Biol. 2013 Sep;166(1):99-108. doi: 10.1016/j.cbpb.2013.07.007. Epub 2013 Jul 20. Comp Biochem Physiol B Biochem Mol Biol. 2013. PMID: 23876386 Free PMC article. - Biological age is a novel biomarker to predict stroke recurrence.
Soriano-Tárraga C, Lazcano U, Jiménez-Conde J, Ois A, Cuadrado-Godia E, Giralt-Steinhauer E, Rodríguez-Campello A, Gomez-Gonzalez A, Avellaneda-Gómez C, Vivanco-Hidalgo RM, Roquer J. Soriano-Tárraga C, et al. J Neurol. 2021 Jan;268(1):285-292. doi: 10.1007/s00415-020-10148-3. Epub 2020 Aug 12. J Neurol. 2021. PMID: 32789606 - Intra-Monozygotic Twin Pair Discordance and Longitudinal Variation of Whole-Genome Scale DNA Methylation in Adults.
Zhang N, Zhao S, Zhang SH, Chen J, Lu D, Shen M, Li C. Zhang N, et al. PLoS One. 2015 Aug 6;10(8):e0135022. doi: 10.1371/journal.pone.0135022. eCollection 2015. PLoS One. 2015. PMID: 26248206 Free PMC article.
References
- Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. - PubMed
Publication types
MeSH terms
Grants and funding
- P30 ES000002/ES/NIEHS NIH HHS/United States
- R01 ES015172/ES/NIEHS NIH HHS/United States
- R01 ES015172-01/ES/NIEHS NIH HHS/United States
- ES00002/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical